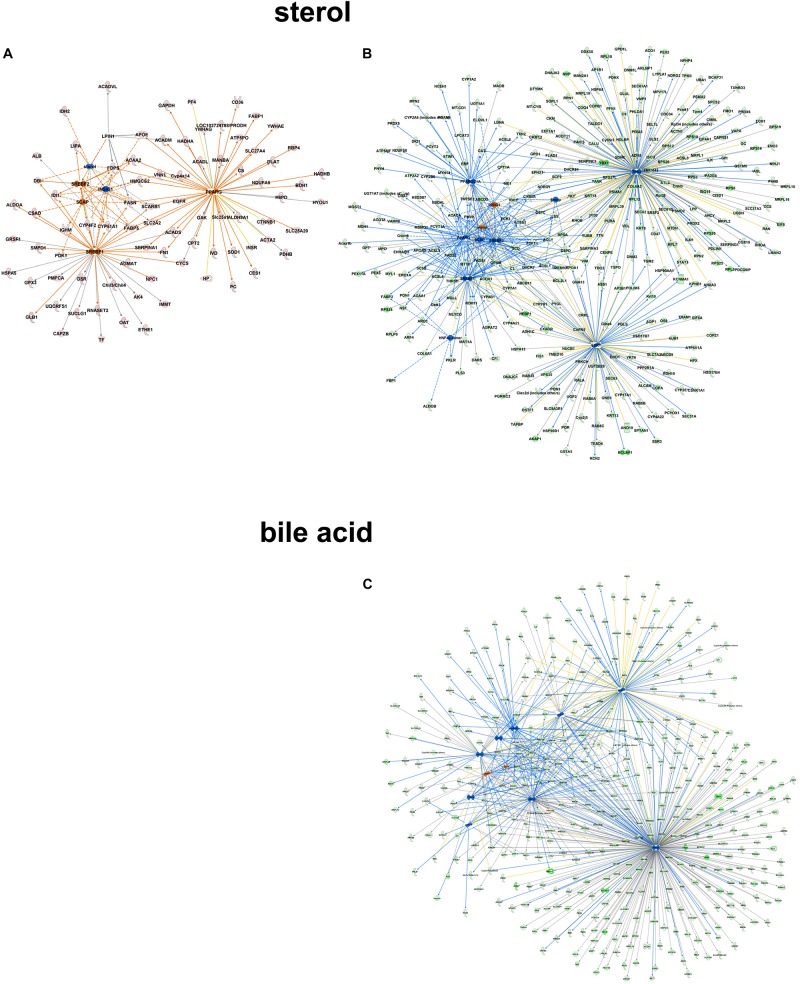
FIGURE 9.
Upstream regulator metabolites in peroxisomal proteins. All proteins identified in peroxisomal fractions of C57Bl6 mice and alb-SREBP-1c mice were separated, according to overrepresentation in the genotypes, and subjected to IPA® Core analyses. Upstream regulator molecule networks are shown for sterol in C57Bl6 overrepresented proteins (A), or in alb-SREBP-1c overrepresented proteins (B), and for bile acid in alb-SREBP-1c overrepresented proteins (C). Upstream regulator molecule networks are shown for sterol (A) and bile acid (B). Hub proteins for sterol in C57Bl6 were PPARG, SCAP, LPIN1, INSIG1, SREBF1, and SREBF2. Hub proteins for sterol in alb-SREBP-1c were: PPARG, SCAP, INSIG1, SREBF1, SREBF2, HNF4α dimer, ESR1, TP53, and PPARGC1A. Hub proteins for bile acid in alb-SREBP-1c were: PPARA, FOXO1, FGF19, NR1H4, NR0B2, HNF4A, MLXIPL, NR5A2, FOXA2, RXRA, HNF1A, and PPARGC1A. Color code, according to IPA® analyses, for molecules: green: overrepresented in alb-SREBP-1c (negative fold change in dataset); red: overrepresented in C57Bl6 (positive fold change in dataset), and for arrows: yellow: findings inconsistent with the state of the downstream molecule; blue: inhibition, consistent with the state of the downstream molecule; orange: increase, consistent with the state of the downstream molecule. Solid arrows indicate a direct interaction, and dotted arrows an indirect interaction, of connected molecules.