Figure 1.
Transcriptome Changes under Oxidative Stress
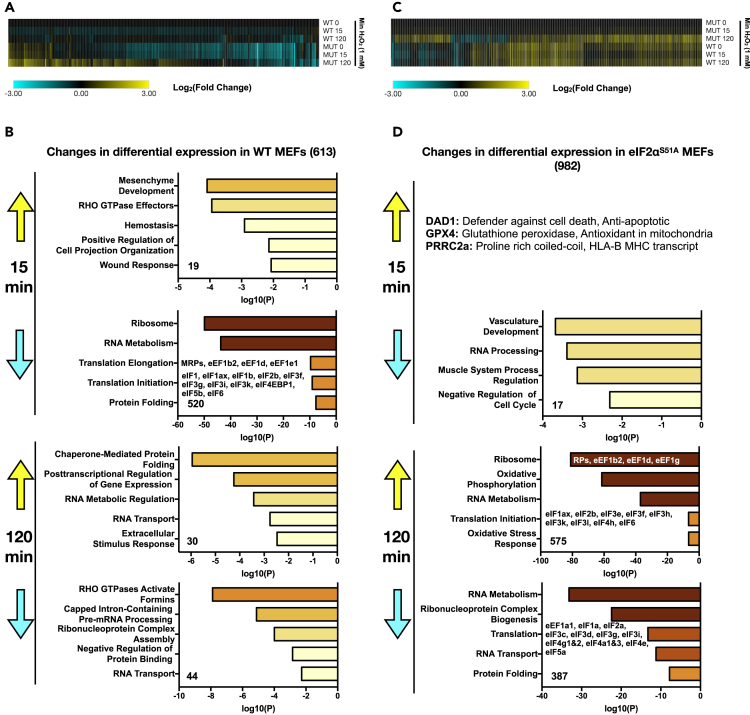
(A) WT and eIF2αS51A MEFs were either untreated or treated with H2O2 (500 μM) for 15 or 120 min. Total RNA was extracted and analyzed by RNA-seq. The data represent a compilation of significant changes in expression (p ≤ 0.05) found in WT MEFs at either the 15- or 120-min time point. Results are normalized to untreated WT MEFs at time 0 and displayed as a clustered heatmap. Upregulated transcripts are shown in yellow and downregulated transcripts in blue. See Data S1 and S2 for complete list (GEO: GSE137409).
(B) Individual list of H2O2-induced and H2O2-repressed mRNAs (±3-fold change, p ≤ 0.05) in WT MEFs were loaded into Metascape, and the pathways enriched are indicated at each time point. Colored boxes represent either upregulated (yellow) or downregulated (blue) pathways. See Data S1 and S2 for a complete list (GEO accession number GSE137409).
(C) WT and eIF2αS51A MEFs were either untreated or treated with H2O2 (500 μM) for 15 or 120 min. Total RNA was extracted and analyzed by RNA-seq. The data represent a compilation of significant changes in expression (p ≤ 0.05) found in eIF2αS51A MEFs at either the 15- or 120-min time point. Results are normalized to untreated eIF2αS51A MEFs at time 0 and displayed as a clustered heatmap. Upregulated transcripts are shown in yellow and downregulated transcripts in blue. See Data S1 and S2 for a complete list.
(D) Individual list of H2O2-induced and H2O2-repressed mRNAs (±3-fold change, p ≤ 0.05) in eIF2αS51A MEFs were loaded into Metascape, and the pathways enriched are indicated at each time point. Colored boxes represent either upregulated (yellow) or downregulated (blue) pathways. See Data S1 and S2 for a complete list.