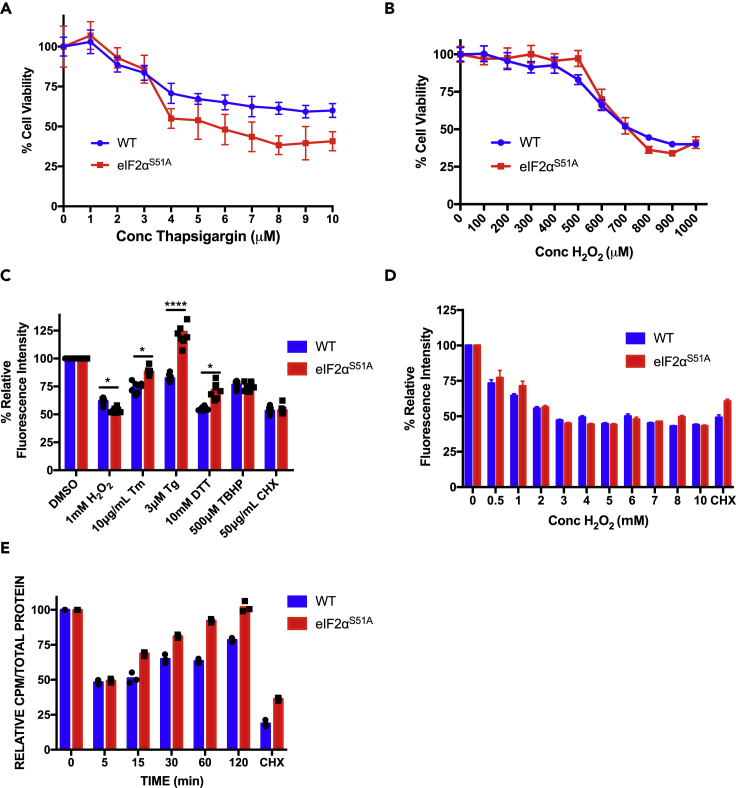
Figure 2.
Effect of Hydrogen Peroxide on Viability and Protein Synthesis
(A) Wild-type and eIF2αS51A mutant MEFs were treated with increasing doses (1–10 μM) of the ER stress inducer thapsigargin for 2 h. MTT assay was used to assess cell viability; see Figure S2A for verified results. The graph represents the mean ± standard deviations of 8 replicates.
(B) Wild-type and eIF2αS51A mutant MEFs were treated with the indicated increasing doses (100–1000 μM) of the oxidative stress inducer hydrogen peroxide (H2O2) for 2 h. MTT assay was used to assess cell viability, see Figure S2B for verified results. The graph represents the mean ± standard deviations of eight replicates.
(C) Wild-type and eIF2αS51A mutant MEFs were treated with the indicated doses of specific stress inducers: H2O2, tunicamycin (Tm), thapsigargin (Tg), dithiothreitol (DTT), and tert-butyl hydroperoxide (TBHP). Cells were treated for 2 h except in the case of tunicamycin, which was added for 4 h. The protein synthesis inhibitor cycloheximide (CHX) was used as a control. Protein synthesis was measured by fluorescently tagging nascent polypeptides and quantifying the fluorescence intensity. The graph represents the normalized mean ± standard deviations of eight replicates. The individual symbols represent individual data points. *p value H2O2 = 0.016, p value Tm = 0.017, p value Tg = 8 × 10−6, p value DTT = 0.013. Statistical significance was determined by t test.
(D) Wild-type and eIF2αS51A mutant MEFs were treated with increasing doses (0.5–10 mM) of H2O2 for 1 h. Protein synthesis was measured with a fluorescent tagging assay. CHX, 50 μg/mL, was added for 1 h as a control. The graph represents the normalized mean ± standard deviations of eight replicates.
(E) Wild-type and eIF2αS51A mutant MEFs were treated with 500 μM H2O2 for increasing periods (5–120 min). Cells were simultaneously labeled with [35S]-methionine for 5 min. Protein synthesis was measured by quantifying radioactive counts per minute (CPM) and adjusted according to the total amount of protein. The graph represents the normalized mean ± standard deviations of at least two independent experiments each done in triplicates. The individual symbols represent individual data points.