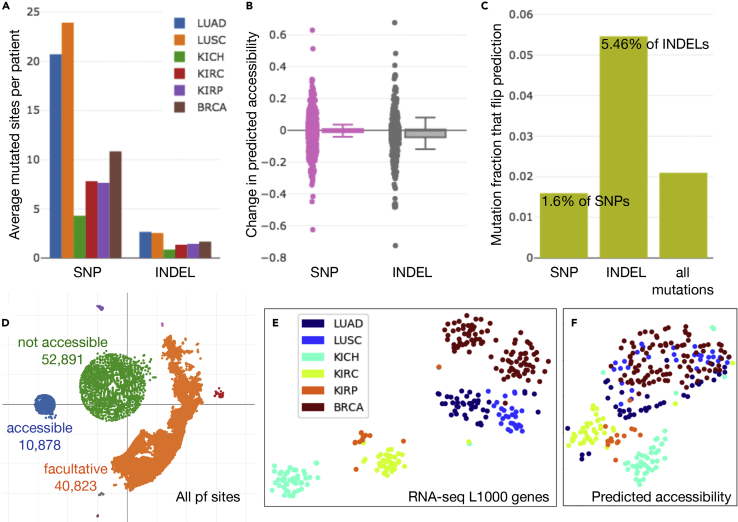
Figure 3.
SNP and INDEL Mutations and Predicted Accessibility Landscape in Tumors
(A) The average number of SNP and insertion or deletion (INDEL) mutations that overlap prediction sites per patient across six TCGA cohorts is shown.
(B) When predictions at sites with mutations were compared with and without applying mutations to the input DNA sequence, the change in predicted accessibility exhibited a higher variance for INDELs than SNPs.
(C) In addition, a larger fraction of sites with INDELs were responsible for a change in the classification decision (flipped prediction) than the fraction of sites with SNPs.
(D) Using t-SNE (perplexity = 50) to visualize the predicted accessibility of individual promoter flank (pf) sites across our selected TCGA samples, we identified which sites were facultative (orange), constitutively accessible (blue), and constitutively not accessible (green).
(E and F) (E) Finally, t-SNE applied to patient samples exhibited different relationships (such as a clear split in BRCA samples) when based on RNA-seq gene expression of the L1000 gene set, than (F) when based on predicted accessibility at all pf sites within each sample (in which case lung and breast cancers appeared to share some common characteristics).