Figure 4.
Promoter and Promoter Flank Accessibility and Checkpoint Gene Expression in LUAD WGS Samples Only and Augmented with Non-WGS Samples
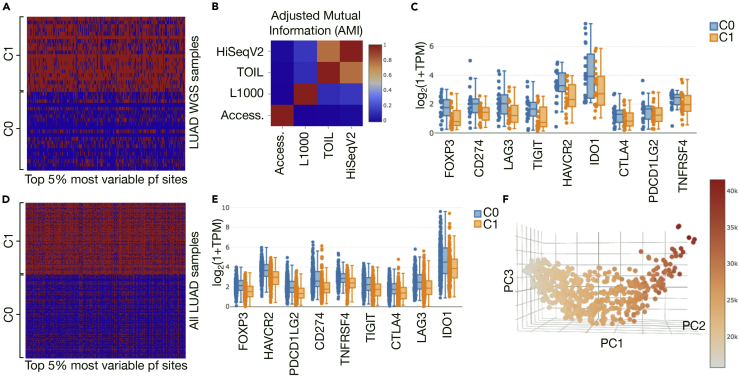
(A) The heatmap and patient sample cluster assignment based on the top 5% most variable promoter and promoter flank (pf) accessibility sites across LUAD samples with WGS available are shown. Cluster 0 (C0) has lower overall accessibility (blue = not accessible), and cluster 1 (C1) exhibits generally higher accessibility (red = accessible).
(B) Adjusted mutual information (AMI) (1) between label assignments based on different data shows higher values (red) between different RNA-seq cluster assignments and low values (blue) between accessibility (Access.) and clusters based on any other data type.
(C) Distribution of key checkpoint gene expression levels (with x axis sorted by significance of two-sided t test between C0 and C1) shows that the low-accessibility group tends to have higher checkpoint levels.
(D) Applying the same procedure to the full LUAD cohort, which also includes predictions for all non-WGS samples, we see a similar split into low- (C0) and high (C1)-accessibility groups.
(E) The same trend in checkpoint expression is observed, with FOXP3 again appearing as the most significant difference (two-sided t test with Benjamini-Hochberg adjusted p = 4.53 × 10−19).
(F) Plotting promoter and flank accessibility with respect to its first three principal components (PC1–3) and coloring points by total number of accessible sites in a sample reveals a smoothly varying relationship, motivating a correlation-based approach to exploring the relationship between overall accessibility and gene expression levels.