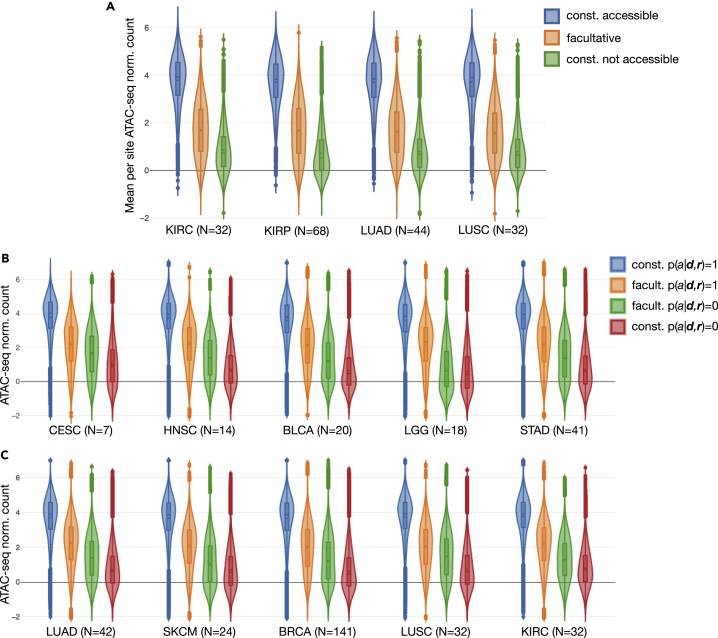
Figure 7.
Validation of Our Promoter and Promoter Flank DNA Accessibility Predictions in TCGA with Empirical ATAC-Seq Measurements
(A) The top violin plots show the distributions of per ATAC-seq peak means of normalized counts in lung and kidney cohorts, for sites we labeled as constitutively (const.) accessible, facultative, or const. not accessible (based on our analysis shown in Figure 3D). Peak count values along all y axes were log transformed and quantile normalized as provided by the authors of the empirical study.
(B and C) Distributions of ATAC-seq peak normalized counts for all prediction sites across all available samples were further broken down per cohort by classification decision (accessible, , and not accessible, ) in addition to site category. Site categories were either facultative (facult.) or constitutive (const.), the latter including both const. accessible as well as const. not accessible. The number of TCGA samples that contributed to each plot is shown (N = ). (B and C) Only TCGA samples for which we had made predictions and were also empirically measured were used, but (A) utilized all available measured samples. The distribution plots were informed by N * 61,342 data points in (B and C), whereas for (A), where we considered the mean value for each site, there were only 61,342 data points total within each cohort.