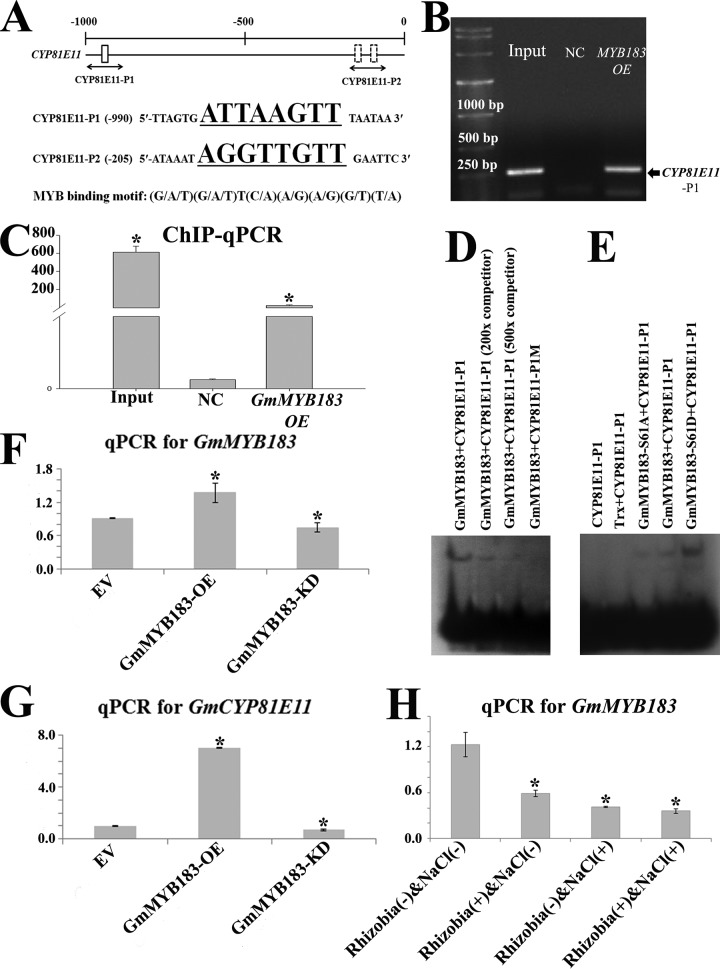
Fig. 3.
GmMYB183 binds to a MYB-specific Cis-element present in the promoter of GmCYP81E11. A, Distribution of MYB binding motifs (G/A/T)(G/A/T)T(C/A)(A/G)(A/G)(G/T)(T/A) in the promoter of GmCYP81E11. Position of the probe (underlined sequence) used for ChIP-based binding assay is shown below the gene. Position Weight Matrix of MYB binding motifs was curated in the JASPAR database (http://jaspar.genereg.net/cgi-bin/jaspar_db.pl?rm=browse&db=core&tax_group=plants). B, and C, ChIP-qPCR assays indicates that GmMYB183 binds to the promoter sections containing the MYB-binding motifs in vivo. The empty vector was used as a negative control (NC). An asterisk indicates a significant difference (p ≤ 0.05) to the NC according to Student's t test. FC means fold change. D, and E, EMSA assays showed that GmMYB183 specially binds to the GmCYP81E11-P1 fragment from the GmCYP81E11 promoter (D), and phosphorylation at S61 of GmMYB183 enhances this interaction (E). GmCYP81E11-P1 (ttttATGTATTAGTGATTAAGTTTAATAACGTGA) or a mutated version with its ATTAAGTT core sequence changed to ATAAAGTT (GmCYP81E11-P1M) was labeled as a probe, and 200 or 500 folds of unlabeled double strand GmCYP81E115-P1 fragment was set as the competitor. F, and G, The transcription levels of GmMYB183 (F) and GmCYP81E11 (G) in soybean transgenic roots expressing empty vector (EV), GmMYB183-overexpression (GmMYB183-OE) and RNAi (GmMYB183-KD) constructs, respectively. Data presented are mean ± S.E. (n = 3). H, Transcription of GmMYB183 in soybean roots treated with R(-)Na(-), R(+)Na(-), R(-)Na(+) and R(+)Na(+). Data represent mean values ± S.E., each sample was analyzed with three biological replicates. An asterisk indicates a significant difference (p ≤ 0.05, Student's t test) between treatment [R(+)Na(-), R(-)Na(+) and R(+)Na(+)] and the control [R(-)Na(-)].