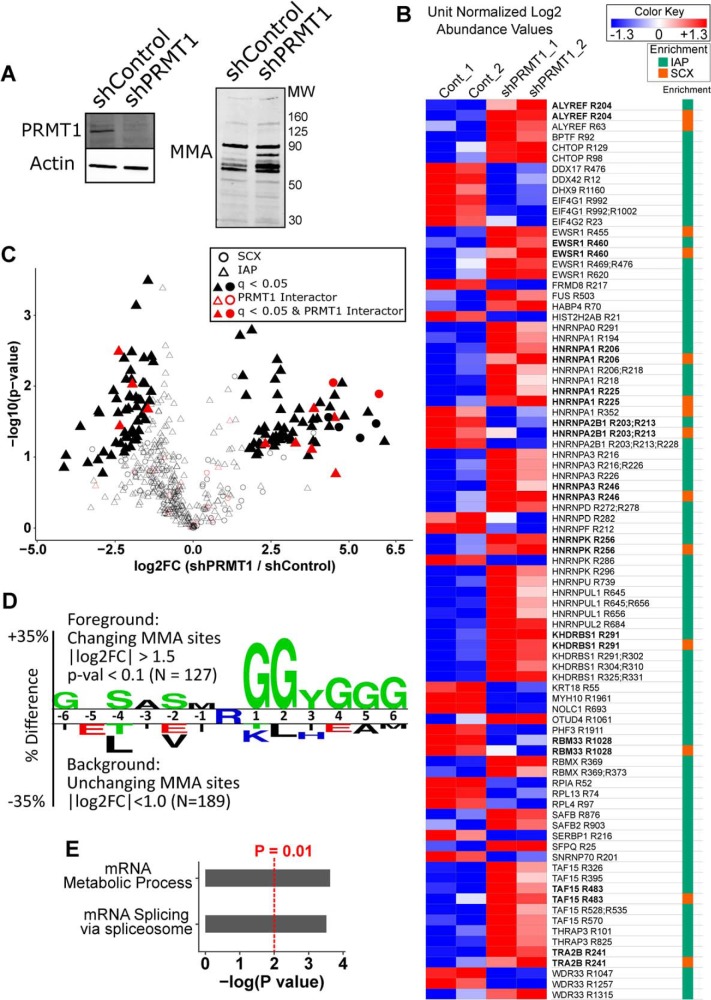
Fig. 3.
Quantitative Analysis of MMA peptides from shPRMT1 293T cells. A, Western blotting confirmed reduced PRMT1 expression and increased MMA levels upon PRMT1 knockdown. 293T cells expressing shControl or shPRMT1 were lysed and analyzed by Western blotting with antibodies against either PRMT1 or MMA. Actin was used as an equal loading control. B, The MMA methylome is substantially altered by PRMT1 knockdown. Heatmap of peptide level differences for methyl peptides captured by SCX and IAP, sorted by gene name. Median normalized log2 LFQ values were unit normalized and colored by fold change as indicated. Methyl peptides measured by both techniques are bolded and showed good quantitative agreement. The column at right indicates which methyl peptide enrichment protocol identified each peptide (SCX or MMA IAP). Bold text indicates peptides identified in both SCX and MMA IAP. C, Volcano plot of MMA peptides enriched by SCX and IAP demonstrating 61 and 58 significantly increased and decreased methyl peptides, respectively, in PRMT1 knockdown cells. The shape indicates which methyl peptide enrichment protocol identified each peptide. Filled shapes indicate q-value < 0.05 by permutation t test in Perseus. Red points denote known interactors of PRMT1 according to the EBI Int Act database (60), and significantly changing MMA peptides were enriched for PRMT1 interactors (p = 0.044 by Fisher's Exact test). D, Two sample motif analysis of changing MMA sites recapitulated the known RGG motif of protein arginine methylation. The motif was generated using Two Sample Logo by comparing MMA peptides with absolute value of log2 fold change > 1.5 against MMA peptides with absolute value of log2 fold change < 1. A p value of 0.05 was used to generate the motif. E, Gene ontology analysis of MMA peptides with absolute value of log2 fold change > 1.5 against MMA peptides with absolute value of log2 fold change < 1D) demonstrated that changing MMA sites were enriched for proteins with mRNA metabolic process (GO:0016071) and mRNA splicing (GO:0000398). Enriched ontologies were identified using GOrilla and passed an FDR q-value < 0.25.