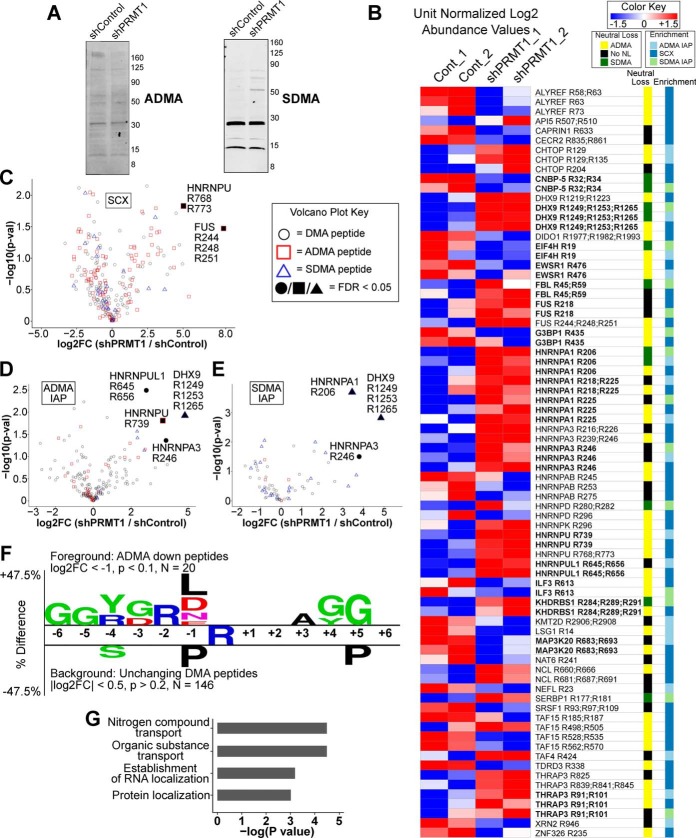
Fig. 5.
Quantitative Analysis of DMA peptides from shPRMT1 293T cells. A, Immunoblot of ADMA and SDMA on 293T cells expressing shControl or shPRMT1. 293T cells expressing shControl or shPRMT1 were lysed and analyzed by Western blotting with antibodies against either ADMA or SDMA. ADMA levels were decreased, and SDMA levels were increased in PRMT1 knockdown cells. Actin was used as an equal loading control and is shown in Fig. 3A. B, The DMA methylome is substantially altered by PRMT1 knockdown. Heatmap of dimethyl peptide levels enriched by SCX, ADMA IAP, or SDMA IAP, sorted by gene name. Median normalized log2 LFQ values were unit normalized and colored by fold change as indicated. The columns at right indicate observed neutral losses (ADMA or SDMA) and which methyl peptide enrichment protocol identified each peptide. Bold text indicates peptides that were identified by multiple enrichment protocols. C–E, Volcano plots of DMA peptides enriched by SCX (C), ADMA IAP (D), and SDMA IAP (E) demonstrating significantly increased DMA peptides in PRMT1 knockdown cells. The type of neutral loss, if observed, is indicated by shape, and the filled in points indicate q-value < 0.05 by permutation t test in Perseus. F, Two-sample motif analysis of downregulated dimethyl peptides showing ADMA neutral loss compared with unchanging background dimethyl peptides. All downregulated ADMA peptides from all experiments were combined for the foreground with log2 fold change < −1 and a p value < 0.1. All unchanging DMA peptides from all experiments were used as the background with a log2 fold change between 0.5 and −0.5 with a p value > 0.20. G, Gene ontology analysis revealed that proteins with significantly changing ADMA sites were enriched for nitrogen compound transport, organic substance transport, establishment of RNA localization, and protein localization. ADMA peptides with absolute value log2 fold change > 1 and Student's t test p value < 0.1 were compared against ADMA peptides with absolute value log2 fold change < 0.5 and Student's t test p value > 0.2. Enriched ontologies were identified in GOrilla and passed an FDR q-value < 0.25.