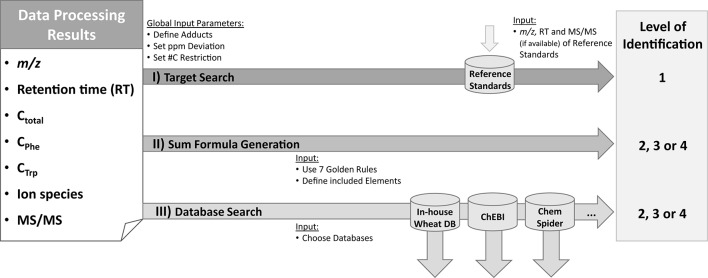
Figure 2.
Identification and annotation workflow. Based on the information from the raw data [m/z and retention time (RT), availability of MS/MS spectra] as well as from MetExtract II evaluation [#C in total (Ctotal); from Phe-TL (CPhe) or Trp-TL (CTrp); ion species] three steps are performed for each detected metabolite. For target search, accurate m/z and RT of the metabolite are compared with authentic reference standards measured in the same sequence and the same analytical method. This step can result in level 1 identification. In the second step, sum formulas are generated using the Seven Golden Rules algorithm (Kind and Fiehn, 2007). The third step consists of database search based on the m/z and the known number of carbon atoms for each metabolite. Identification levels are based on the Metabolomics Standards Initiative (Blaženović et al., 2018). ChEBI database (Hastings et al., 2013); ChemSpider database (Pence and Williams, 2010)