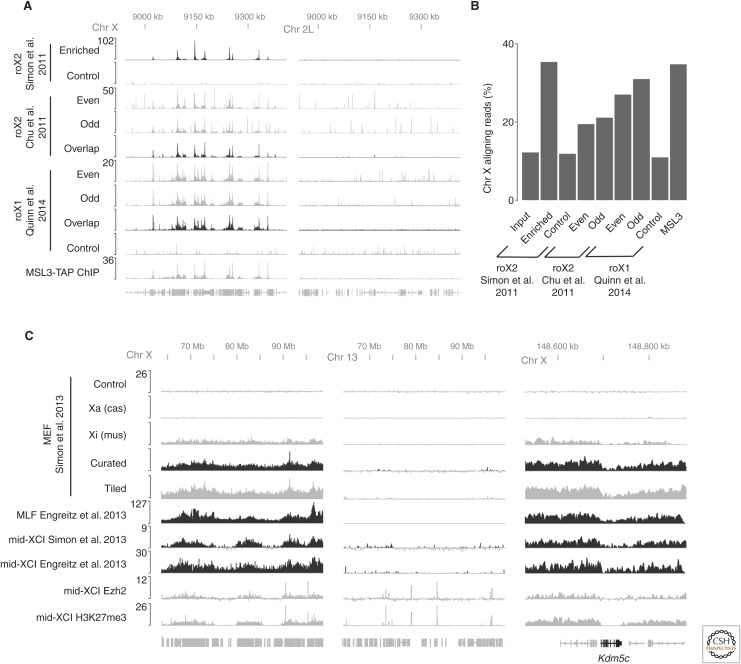
Figure 3.
Comparison of different hybridization capture experiments. (A) roX RNAs enrichment on Chr X is highly reproducible among several published studies and agrees with ChIP experiments targeting subunits of the male specific lethal (MSL) complex (Alekseyenko et al. 2008). The roX1 profiles are from a fusion experiment that uses formaldehyde cross-linking and formamide hybridization buffers (Quinn et al. 2014). Autosomal data (Chr 2L) show how controls and postprocessing (overlap) can help to bioinformatically reduce off-target hybridization signals from odd and even oligonucleotide pools. (B) Bar graph illustrating the fraction of purified DNA from different enrichment strategies that originates from Chr X. (C) Xist long noncoding RNA (lncRNA) coats broad regions of inactive Chr X (Xi) except for several escapee genes including Kdm5c (right panel). Signals from tiled oligonucleotides and curated oligonucleotides agree well (Simon et al. 2013). Xist enrichment on chromosome X was found to highly correlate with PRC2 (Ezh2 ChIP; Pinter et al. 2012) and H3K27me3 ChIP (Pinter et al. 2012) during X-chromosome inactivation (XCI). Hybrid cell lines in which active (Xa) and inactive (Xi) X chromosomes can be distinguished based on their allele-specific differences validate that signals arise from biological cross-linking and not artifact (Simon et al. 2013).