Abstract
Genomic data offer valuable insights that can be used to help find treatments and cures for disease. Precision medicine, defined by the NIH as “an emerging approach for disease treatment and prevention that takes into account individual variability in genes, environment, and lifestyle for each person,” is gaining acceptance among physicians, who are beginning to integrate patient-centric data analysis into clinical decision-making. Although precision medicine makes use of various types of data, this piece focuses on molecular characterization data specifically, as the discoveries yielded from these data can advance thinking around clinical care for cancer patients. Our pediatrics genomics team at the University of California Santa Cruz Genomics Institute is uniquely situated to discuss the use of shared genomic data for clinical benefit because our collaborations with hospital partners in the United States and internationally rely on big-data comparative genomic analysis. Using shared data, Treehouse Childhood Cancer Initiative develops methods for comparative analysis of tumor RNA sequencing profiles from single patients for the purposes of identifying overexpressed oncogenes that could be targeted by therapies in the clinic. To enable and improve this analysis, we continuously increase the size of our data compendium by adding public pediatric tumor RNA sequencing data sets. We developed an approach for assessing the quality of shared RNA sequencing data to ensure the integrity of the data. In this approach we calculate the number of mapped exonic nonduplicate (MEND) reads, applying a 10 million MEND read minimum threshold for inclusion in our comparative analysis. In collaboration with Stanford University and Lucile Packard Children's Hospital Stanford, our team at Treehouse Childhood Cancer Initiative explores the value to researchers everywhere of shared genomic data for clinical utility and the challenges of data sharing that threaten to impede otherwise rapid advances in precision medicine. This Perspective offers recommendations for maximizing the use of genomic data to make discoveries that will benefit patients.
THE VALUE OF SHARED GENOMIC DATA
Genomic information has offered clues for better diagnosis, prognosis, and treatment of cancers. Although clinical trials have largely defined treatment standards for newly diagnosed cancer, the optimal approach to salvage therapy, used when a patient's disease does not respond to standard therapy or recurs, remains elusive except in limited scenarios in which a cure can reasonably be expected. For patients who face long odds of cure, clinicians must weigh the benefits and downsides of an increasingly wide array of therapeutic options for which there are often limited data, particularly in rare tumor types. Sorting through these options to prioritize the treatments with the highest benefit:toxicity ratio is often an exercise in conjecture rather than science. To address this problem, oncologists are increasingly seeking tumor biomarker information. This is particularly true as the field of genomics grows and the volume of genomic, transcriptomic, and proteomic information expands. As data accrues on the value of molecularly targeted therapy, the need to rapidly identify which tumors express the target, and which patients are most likely to benefit, has become more urgent.
The emergence of genomic medicine, a data-driven discipline, has spurred the biomedical community to examine the potential benefits of genomic data sharing. For example, data sharing initiatives such as the Matchmaker Exchange (https://www.matchmakerexchange.org/) that enable researchers to share and compare DNA sequence variants and associated patient phenotype information have already led to the discovery of novel human disease genes and have become indispensable in rare disease research. Along with genomic medicine as a whole, the cancer genomics community has benefited from data sharing and combined analysis of multiple data sets. In recognition of the value of data sharing, the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists issued a joint statement that strongly encourages laboratories to contribute curated somatic variants to public databases to facilitate interpretation (Li et al. 2017).
Pediatric cancer research is uniquely situated to capitalize on the promise of genomic data sharing. Because pediatric cancers are rare and heterogeneous at the molecular level, individual hospitals and research institutions are unlikely to be able to create cohorts that are large enough for statistically meaningful analysis. Several important pediatric cancer genomic databases and portals have emerged to enable the analysis of multiple data sets (Sweet-Cordero and Biegel 2019). In addition, because pediatric cancers harbor fewer mutations than adult cancers and epigenetic aberrations are frequently implicated in pediatric cancer development, functional genomic information is important for clinical decision-making. RNA sequencing data can provide a readout of both genetic and epigenetic changes in the tumor. In a recent evaluation by the FDA, it was determined that relative rather than absolute RNA sequencing–derived transcriptome profiles are robust enough for clinical analysis (Xu et al. 2016). However, the derivation of relative gene expression profiles requires access to large RNA sequencing data sets from both cancer patients and unaffected individuals, making the sharing of RNA sequencing data essential for the implementation of RNA sequencing analysis in the clinic. Privacy concerns are lessened in the cancer genomic space because somatic mutations are not heritable and only characteristic of an individual tumor rather than the patient's family. Sharing of tumor RNA sequencing–derived expression profiles, as in our Treehouse initiative (treehousegenomics.ucsc.edu), is even less restrictive, as such profiles do not contain identifiable sequence information.
THE CHALLENGES OF GENOMIC DATA SHARING AND WAYS TO MOVE FORWARD
Researchers who seek to advance treatment and cures for patients through the use of shared data face challenges that are systemic, technical, and educational.
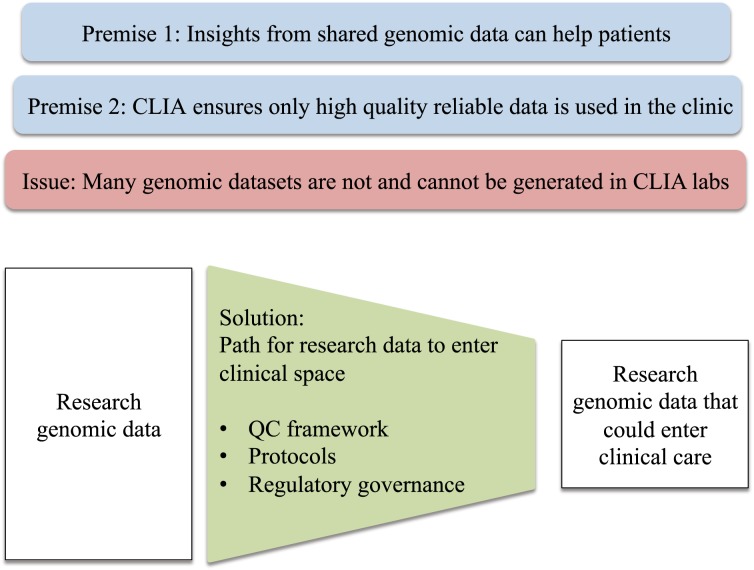
One significant challenge is the lack of a clear path for genomic data sets collected in research settings to enter clinical care (Fig. 1). Compliance to clinical laboratory standards set by Clinical Laboratory Improvement Amendments (CLIA) and regulatory approval by the FDA are commonly accepted routes to for gaining authorization to use genomic testing to inform treatment (Shevchenko and Bale 2016). However, the existing framework is not focused on research-derived data.
Figure 1.
A key issue that hinders the realization of the full potential of genomic data is the lack of a clear path for the data generated in research settings to enter clinical care.
Many multigene DNA sequencing tests, aimed at studying the DNA sequence of patient cells, are administered as CLIA-approved laboratory developed tests (LDTs), and several have achieved approval by the FDA (https://www.fda.gov/news). This precedent makes it easier for other institutions to develop similar tests or adopt existing assays cleared by the FDA. However, for other types of genomic data, such as transcriptome sequencing data, there are few LDTs available and the path to FDA approval is less clear. In addition, much of the cancer genomic data has been collected as part of research rather than clinical efforts, making its integration into a traditional CLIA framework challenging (Fig. 1). Although adherence to sound protocols helps to ensure uniform quality and the reliability of shared genomic information, the existing CLIA framework could be reconsidered in light of the precision medicine applications that aim to provide personalized rapidly evolving biological and computational genomic analyses. The goal must be to drive forward discoveries that are translated quickly to benefit patients, regardless of whether the underlying data were studied in a research or clinical context. To maximize the benefit for patients in need of immediate care, particularly those with rare diseases, the CLIA framework should be sufficiently flexible while maintaining rigorous standards.
Although funding agencies, academic and medical institutions, and researchers are increasingly recognizing the value that shared data brings to medical discovery, adoption of consistent data standards, particularly with respect to sequencing protocols and metadata annotation, lag behind. As a consequence, genomic data generated by outside institutions is often difficult to find, access, and use. The absence of data annotation standards results in confusion on data elements critical to use and analysis, such as disease designation (Learned et al. 2019). Although there are internationally accepted World Health Organization International Classification of Diseases standards for the diagnostic classification of cancer types and anatomic locations, this system is insufficiently granular, quickly outdated, and not utilized by many institutions, limiting harmonization. To address this, initiatives such as American Society for Clinical Oncology's mCODE (ASCO 2019) are developing structured data elements for oncology health records. Similarly, although guidelines for DNA variant quality, pathogenicity, and potential impact on the patient exist thanks to the American College of Medical Genetics (among others), quality standards for transcriptomic and proteomic data and other types of genomic data do not. Uneven quality limits the ability of researchers to rely on that data to investigate molecular signatures of disease. Similarly, the library preparation is conducted without the benefit of universally adopted protocols. For example: a ribosomal deplete, polyadenylation (poly(A)) or exome capture protocol may be used in RNA-seq library preparation; these protocols differ in the types of input RNA that could be processed, and as a result, the gene expression measurements are not directly comparable (Bush et al. 2017). Although individual teams like ours have developed methods to measure data quality, to date, no validated bioinformatic method of assessing quality or correcting for these differences that result from preparation methods exists. One solution is to produce standards that must be adhered to in order to meet existing data sharing requirements, thus incorporating an effort to regularize approaches and reporting within the research incentive framework.
Data sharing also is impacted at the earliest stages of data gathering: obtaining patient consent and performing the tumor biopsy. Data sharing requires patient consent, yet consent documents often fail to address the importance of data sharing and its potential risks. Patient consent must expressly recognize the importance of patient data in aiding new discoveries. There is evidence that patients will respond in a way that spurs data sharing: When patients are told about the research value of sharing data, they are disposed to consent to broad-based sharing and use of their data (Richter et al. 2019). For ethical reasons, informed consent for tumor biopsies to obtain tissue for genomic profiling should include a careful review of the potential for clinical benefit. Pro forma DNA mutation tissue testing conducted by CLIA laboratories has been adopted by some institutions, resulting in mandatory transfer of tissue for standard testing at the site, leaving only small amounts of residual tissue for research testing that may lead to new insights both for the patient and, through data sharing, for other patients. It also may impact the type of sequencing that is done. Therefore, the biopsy must be planned considering the kinds of testing envisioned, including planning for possible use of such tissue at a later date, to maximize use as a shared data resource. One solution is to redefine the process for biopsy planning and to engage the immediate care team, the surgeon, and the pathology team at the outset, before surgery, to ensure that the tissue is handled appropriately. This requires a structural shift away from a focus strictly on individual interests toward one that also considers collective interests.
At biopsy, the utility of data sharing is influenced by the amount of tissue resected, how tumor tissue is stored following biopsy, the time between storage and sequencing, the sequencing protocol, and the quality of the tissue sample. Yet often individual decisions in these areas are made without consideration as to data sharing or consideration of patient and family wishes in this regard. Frequently, the tissue preservation step is divorced from the intended use—for example, tissue may be placed in formalin, which impairs sequencing quality and future utility, despite advance notice that the patient or family hopes to have the tissue sequenced or is committed to using the tissue to help advance research efforts. One solution to this would be to consent patients up front on protocols that are designed with data sharing in mind, instead of consenting patients for biobanking followed by a downstream “use protocol.”
Compounding this difficulty is that access to genomic data is often controlled because of concerns that sharing of primary sequence data may result in patient identification or reidentification by future researchers. In these cases, researchers seeking data are often bound to restrictive agreements that limit the dissemination of genomic data within the broader research community. Such agreements create delay and may block data sharing when rules or laws regarding data privacy conflict or choice of law is disputed; additionally, they may need to be regularly updated as study methods are amended. To address institutional concerns about giving data to other institutions, Global Alliance for Genomics and Health, an international coalition tasked with facilitating data sharing, has developed a federated model for genomic data sharing that includes sharing of computational pipelines rather than data itself. Although this technical solution addresses concerns of privacy protection, only institutions with significant resources can afford to pay the computational fees or have the technical know-how required for computing data at the data-holder's site through a dockerized container that operates on a cloud-based platform. It is necessary to reduce the cost of genomic data computation borne by individual researchers through a combination of increased market competition and funder support.
A more rational framework to evaluate privacy and identification risks is needed to minimize increasingly protective barriers to genomic data disclosure while maintaining necessary patient confidentiality. This requires education of policy-makers, institutions, and regulators about the types of genomic data and their associated risks. Although some types of data, such as rare germline data, present legitimate risks of identity disclosure, other types of data, such as transcriptome and functional genomic data, present less risk (Greenbaum et al. 2011). Medical institutions as well as patients themselves require assistance from genomic experts in assessing and understanding risks presented by genomic data. The current risk-averse model results in a thick regulatory fog that hangs over many data requests, often making the process too time-consuming or inscrutable to navigate, particularly for researchers whose focus is on finding disease cures rather than data mining. One solution is to characterize genomic information along a continuum of risk and to develop control guidelines that attach to each risk category. For instance, in our area of expertise, RNA-seq data, the risk of reidentification or privacy violation is de minimis. Therefore, access should not be controlled. In contrast, germline information is high risk, and controlled access is appropriate. Tailoring agreements to only cases of medium to high risk would significantly loosen regulatory burdens and maximize data use.
Similarly, education plays a crucial role in actualizing the translational use of genomic data. Although shared genomic data offers both research and clinical benefits, academic scientists and medical professionals often do not share a language or knowledge base that allows for an easy understanding of how genomic data fits within a clinical decision-making framework. Effective understanding requires additional training across disciplines, in particular for those trained prior to the era of genomic discovery. Studies to evaluate how personalized treatment recommendations generated by genomic profiling impact treatment outcomes are also needed to assist clinicians in prioritizing therapies identified by genomic testing. Reports that summarize the findings of genomic testing must provide sufficient information that the clinician can judge the quality of the analysis and the value of therapeutic targeting of the abnormalities identified, ideally presented in a manner that is easily understandable. In our experience, it is critical that the report nomenclature is adapted to clinical requirements, so that genomic information gathered through shared data is transferred effectively into the clinical environment.
Perhaps most important in reducing barriers to genomic data sharing and galvanizing the design and implementation of standardized data and sequencing procedures is a change in how scientific research is supported. Current support structures favor siloed research teams within large, wealthy institutions; smaller studies with limited resources or focused on deprioritized areas are at a disadvantage with respect to the resources needed for data gathering and analysis. Incentive structures, such as funding, career advancement, or awards that favor the collection and sharing of data as a community resource would advance data sharing. Funding agencies and foundations are ideally placed to lead such change.
Data sharing offers extraordinary potential for clinical benefit. Existing practices and frameworks are adaptable and can integrate information from data available across different research initiatives, leading to valuable clinical insights. In particular, how we disclose and make available data must assume data use by researchers and clinicians from large and small institutions around the world, necessitating standardization, accessibility, and de minimis cost. Similarly, it should be possible to integrate data from multiple research sites into the CLIA framework through rigorous quality control standards, protocols, and supporting regulatory requirements. Recent advances in technology make data sharing obtainable on a broad scale; through regulatory and procedural refinement of existing structures, the discoveries made possible by data sharing will stimulate progress in the clinic.
ADDITIONAL INFORMATION
Acknowledgments
The authors thank the Treehouse Childhood Cancer Initiative team at UC Santa Cruz: Lauren Sanders, Jacob Pfeil, Allison Cheney, Holly Beale, Geoff Lyle, Katrina Learned, Anouk van den Bout, Ellen Kephart, Rob Currie, and Sofie R. Salama.
Funding
The authors acknowledge the generous support of St. Baldrick's Foundation Consortium Award and Emily Beazley Kures for Kids Fund Hero Award, the State of California Initiative to Advance Precision Medicine, Unravel Pediatric Cancer, Team G Childhood Cancer Foundation, Live for Others Foundation, Alex's Lemonade Stand Foundation for Childhood Cancer Research, and Lucile Packard Children's Hospital Stanford. D.H. is a Howard Hughes Medical Institute Investigator.
Competing Interest Statement
Dr. Vaske's spouse is an employee of ImmunityBio Inc and has equity interests in NantHealth.
REFERENCES
- ASCO. 2019. mCODE: creating a set of standard data elements for oncology EHRs. https://www.asco.org/practice-guidelines/cancer-care-initiatives/mcode-creating-set-standard-data-elements-oncology-ehrs (accessed September 2, 2019). [Google Scholar]
- Bush SJ, McCulloch MEB, Summers KM, Hume DA, Clark EL. 2017. Integration of quantitated expression estimates from polyA-selected and rRNA-depleted RNA-seq libraries. BMC Bioinformatics 18: 301 10.1186/s12859-017-1714-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Sboner A, Mu XJ, Gerstein M. 2011. Genomics and privacy: implications of the new reality of closed data for the field. PLoS Comput Biol 7: e1002278 10.1371/journal.pcbi.1002278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned K, Durbin A, Currie R, Kephart ET, Beale HC, Sanders LM, Pfeil J, Goldstein TC, Salama SR, Haussler D, et al. 2019. Barriers to accessing public cancer genomic data. Sci Data 6: 1–7. 10.1038/s41597-019-0096-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, et al. 2017. Standards and guidelines for the interpretation and reporting of sequence variants in cancer. J Mol Diagn 19: 4–23. 10.1016/j.jmoldx.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter G, Borzikowsky C, Lieb W, Schreiber S, Krawczak M, Buyx A. 2019. Patient views on research use of clinical data without consent: legal, but also acceptable? Eur J Hum Genet 27: 841–847. 10.1038/s41431-019-0340-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko Y, Bale S. 2016. Clinical versus research sequencing. Cold Spring Harb Perspect Med 6: a025809 10.1101/cshperspect.a025809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet-Cordero EA, Biegel JA. 2019. The genomic landscape of pediatric cancers: implications for diagnosis and treatment. Science 363: 1170–1175. 10.1126/science.aaw3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Gong B, Wu L, Thakkar S, Hong H, Tong W. 2016. Comprehensive assessments of RNA-seq by the SEQC consortium: FDA-led efforts advance precision medicine. Pharmaceutics 8: E8 10.3390/pharmaceutics8010008 [DOI] [PMC free article] [PubMed] [Google Scholar]