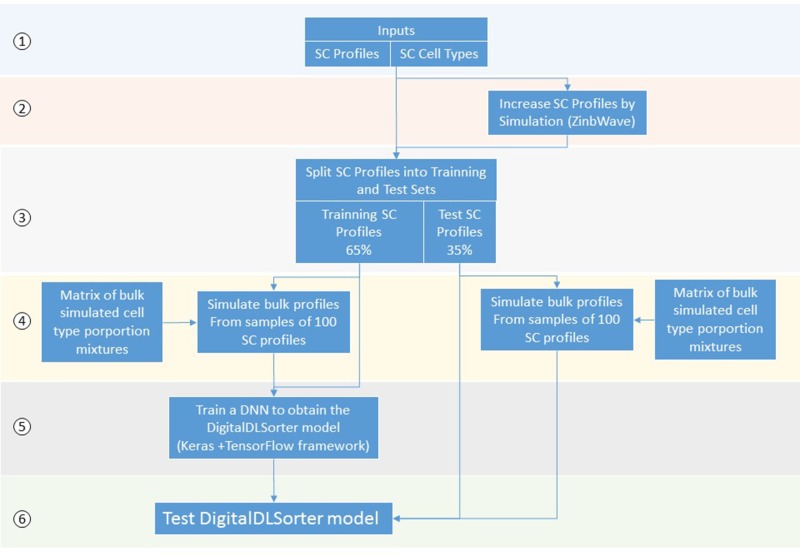
Figure 1.
Scheme of the DigitalDLSorter Pipeline. 1) The pipeline takes a matrix of sc-RNASeq gene expression profiles (SC profiles) and a phenotype file indicating the cell type of each cell. 2) In those experiments were the number of SC profiles for each cell type is low, new SC profiles are simulated based on the real data using ZinbWave framework (Risso et al., 2018). 3) Real and simulated SC profiles are split into training (65%) and test (35%) sets. 4) For each training and test set, bulk samples are created by mixing 100 single cell profiles sampled according to the bulk cell type proportions simulated for each set. 5) A DNN is trained with the training set of bulk profiles together with the corresponding SC profiles. 6) The model obtained is applied to the test set, bulk and SC profiles.