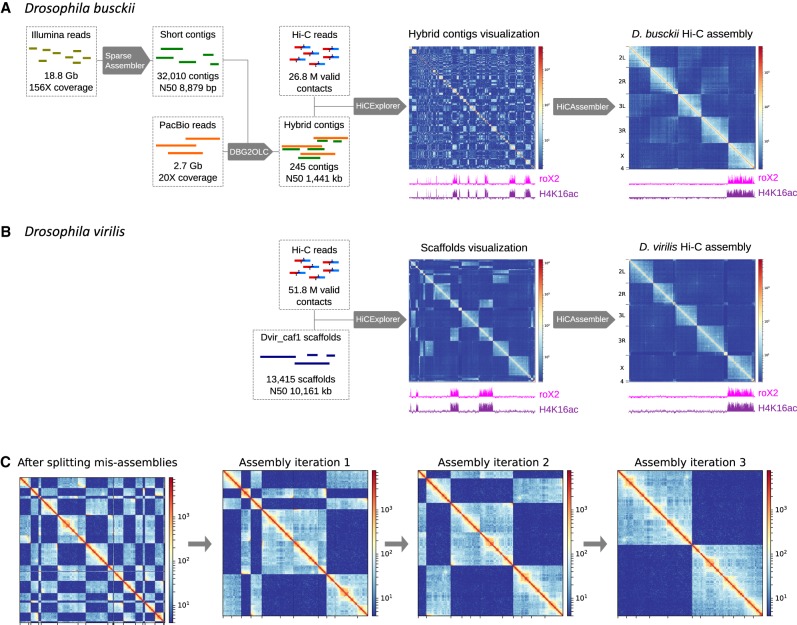
Figure 1.
Hi-C guided chromosome-length assemblies of D. busckii and D. virilis genomes. (A) De novo assembly of D. busckii genome. A hybrid approach integrating long PacBio reads and short contigs assembled from Illumina reads was used to obtain 245 de novo contigs of the D. busckii genome. Assembly of 156× Illumina reads using SparseAssembler (Ye et al. 2012) resulted in 32,010 short contigs. 20× PacBio data was integrated using DBG2OLC (Ye et al. 2016), which increased the N50 more than 100-fold. These 245 hybrid contigs were scaffolded into chromosome-length with Hi-C data using HiCAssembler. Integrity of the X chromosome (identified by whole-genome alignment to D. melanogaster) was validated using ChIRP-seq data of the dosage compensation complex member roX2 (Quinn et al. 2016) and ChIP-seq data of H4K16ac from male D. busckii larvae. (B) D. virilis Hi-C assembly. The existing reference scaffolds of D. virilis (Dvir_caf1 scaffolds) were assembled into full chromosomes using HiCAssembler. The enrichment of roX2 and H4K16ac (male) on one chromosome depicts full integrity of the assembled X chromosome. (C) Overview of HiCAssembler strategy (see Materials and Methods and Supplemental Fig. S3 for a complete description of the algorithm). The figure displays the iterative progression of the Hi-C assembly strategy as in Dudchenko et al. (2017) for a small example Hi-C matrix. First, the original scaffolds are split if they contain misassemblies and small scaffolds are removed. In each iteration of the Hi-C assembly algorithm scaffolds are joined and oriented to form larger and larger Hi-C scaffolds until chromosome-length assemblies are obtained as shown in the last panel where two separated blocks remain. Afterward, the small scaffolds that were initially removed are inserted into the Hi-C scaffolds.