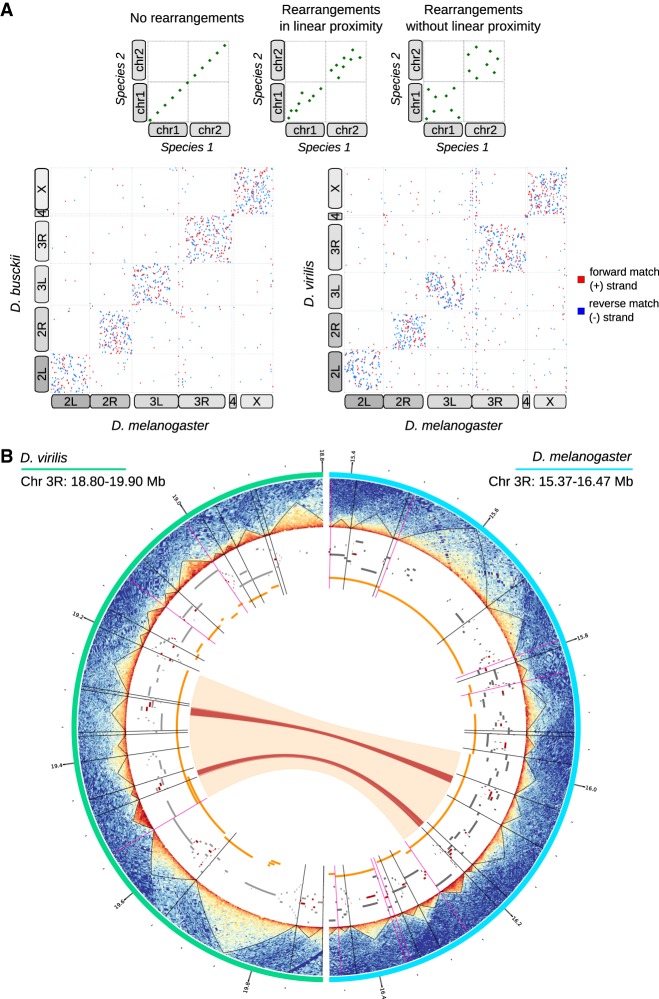
Figure 2.
Extensive genome shuffling during Drosophila evolution. (A, top) Hypothetical whole-genome alignments. If no rearrangements have occurred between two species, whole-genome alignments result in matches that perfectly align at the diagonal (left). If there was a link between linear proximity and synteny breakpoints, matches would be expected to converge near the diagonal (middle). If shuffling happens without linear proximity, matches would occur randomly throughout the whole-chromosome arms (right). (Bottom) Dotplots showing actual whole-genome alignments between D. melanogaster and D. busckii or D. virilis, respectively. Alignments were performed using Mummer4 (Marçais et al. 2018). Forward matches (+ strand) are shown in red; reverse matches (− strand) are displayed in blue. Corresponding chromosome arms are indicated with boxes that are displayed connected if chromosome arms are fused in one species. Karyotypes are additionally depicted in Supplemental Figure S2A. (B) Association between TAD boundaries and synteny block breakpoints. From the exterior to the interior of the Circos plot. (Turquoise) The 18.80–19.90 Mb region of the chromosome 3L in D. virilis, (light blue) the 15.37–16.47 Mb of the chromosome 3L in D. melanogaster, (heatmaps) Hi-C contact heatmaps with TADs displayed as black triangles, (black radial lines) TAD boundaries, (magenta radial lines) TAD boundaries overlapping with synteny block start or end sites, (gray blocks) genes, (red blocks) BUSCOs, (orange blocks) synteny blocks, (orange arc) conserved synteny block between D. virilis and D. melanogaster in the displayed regions, (red arcs) conserved BUSCOs in the displayed regions.