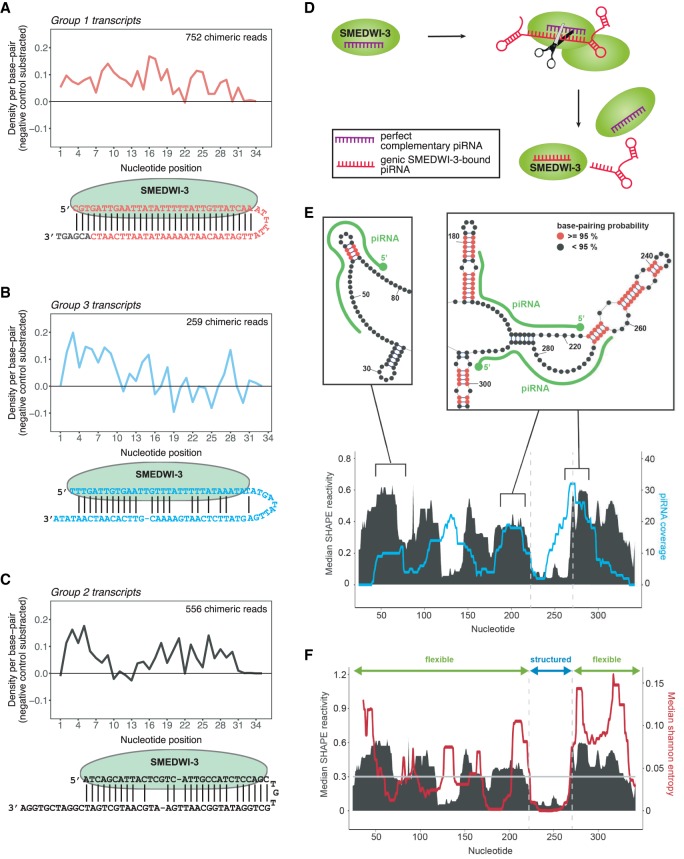
Figure 5.
The base-pairing patterns between SMEDWI-3-bound piRNAs and target mRNAs. (A) Average density of base-pairing events per nucleotide position of the piRNA parts of chimeric reads. Chimeric piRNA parts were mapped onto targeted mRNA fragments in the vicinity of ±20 nt from the mRNA fragment midpoint. Only unique chimeric reads mapping to transcripts of Group 1 were included from all nine replicates. Random piRNA mapping densities were subtracted as negative control. (Below) An illustration of one exemplary chimeric read from group 1 transcripts is shown. The extended mRNA part of the chimeric read is shown in gray. (B) Same as in A for group 3 SMEDWI-3 targets. (C) Same as in A for group 2 SMEDWI-3 targets. (D) SMEDWI-3 degrades a set of mRNAs in a homotypical ping-pong cycle. It recognizes these transcripts with a high degree of complementarity between the SMEDWI-3-bound piRNA and the target mRNA. (E) Plot showing the correlation between experimentally determined median SHAPE reactivity (in black) and the mapping profile of unique predicted antisense piRNAs (in cyan) for histone H2B (SMESG000052758.1). For piRNA mapping, one mismatch was allowed aside from a mandatory seed match for nucleotides 2–8. Exemplary regions from the SHAPE reactivity-constrained secondary structure model are shown above the plots. Double-stranded regions with base-pairing probabilities >0.95 are colored in red. piRNAs (green) are drawn along the structures. (F) Plot showing the correlation between median SHAPE reactivity (in black) and median Shannon entropy (in red). Transcript regions with median SHAPE reactivity <0.3 and a Shannon entropy of <0.04 were considered structured.