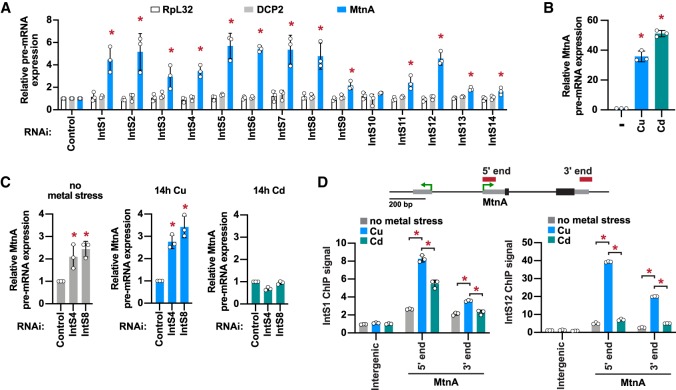
Figure 2.
The Integrator complex is present at the MtnA locus during copper stress and attenuates MtnA transcription. (A) DL1 cells were treated with dsRNAs for 3 d to induce RNAi and depletion of the indicated factors. A total of 500 µM CuSO4 was added for the last 14 h. RT-qPCR was then used to measure the pre-mRNA levels of endogenous RpL32, DCP2, and MtnA. Primer pairs that span an intron–exon boundary were used to specifically amplify pre-mRNAs. Data were normalized to RpL32 mRNA expression and are shown as mean ± SD, N = 3. (*) P < 0.05. (B) DL1 cells were unstressed (–) or treated with 500 µM CuSO4 or 50 µM CdCl2 for 14 h, and RT-qPCR was then used to measure MtnA pre-mRNA levels. Data were normalized to RpL32 mRNA expression and are shown as mean ± SD, N = 3. (*) P < 0.05. (C) DL1 cells were treated with dsRNAs for 3 d and 500 µM CuSO4 or 50 µM CdCl2 was added for the final 14 h, as indicated. RT-qPCR was then used to measure MtnA pre-mRNA levels. Data were normalized to RpL32 mRNA expression and are shown as mean ± SD, N = 3. (*) P < 0.05. (D) The MtnA locus with the locations of ChIP amplicons. Recruitment of IntS1 and IntS12 in unstressed cells (gray) or after the cells had been treated with CuSO4 (blue) or CdCl2 (green) for 14 h was measured using ChIP-qPCR. Data are shown as fold change relative to the IgG control (mean ± SD, N = 3). (*) P < 0.05.