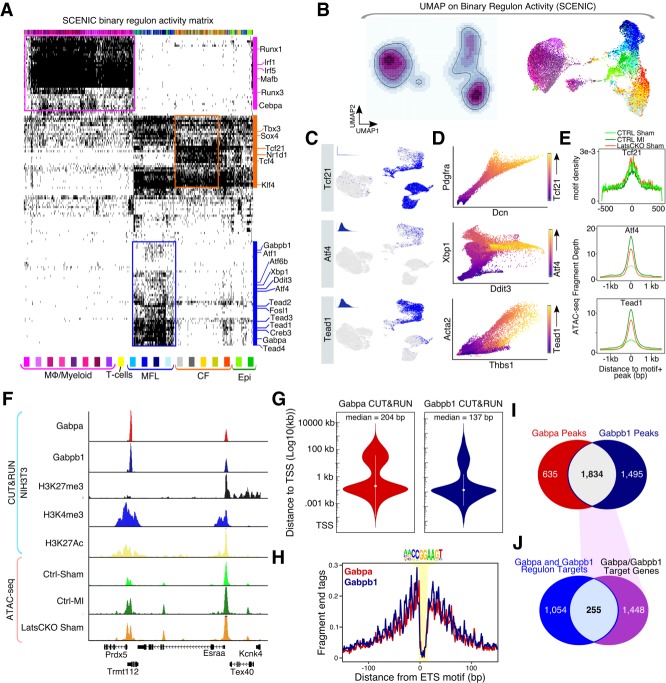
Figure 3.
Tead and ER stress-response-associated transcription factor GRNs are active in myofibroblasts. (A) Binary regulon activity matrix results from SCENIC algorithm carried out on cardiac Drop-seq data. Individual cells are columns, and regulons are rows. (B) Cardiac cell identity UMAP generated from the binary regulon activity matrix. (Left) Regulon activity density plot embedded on UMAP. Light color indicates low cumulative regulon activity, and darker color indicates high cumulative regulon activity. (Right) Binary regulon activity UMAP colored by individual cell cluster identity. (C) Regulon activity for individual cells embedded on the expression based UMAP. Blue highlights active regulon for indicated transcription factor, and gray indicates that a regulon is not active. Insets show the AUCell score histogram for the regulon. (D) MAGIC scatterplots of gene–gene relationships for regulon components. Transcription factor expression encoded by highlighted color gradient. (E) Evaluation of regulon activity using Fast-ATAC. (Top) Global Tcf21 motif enrichment across peaks from each indicated experimental condition. (Bottom) Fast-ATAC read enrichment across motif-containing peaks annotated to genes in either the Atf4 (middle) or Tead1 (bottom) regulons identified in Figure 3A. (F) Genome browser tracks displaying GABP CUT&RUN. (G) Violin plot showing the absolute distance of Gabpa and Gabpb1 peaks to the nearest transcription start site (TSS). (H) CUT&RUN footprint analysis for GABP across the ETS motif. (I) Venn diagram showing intersect of Gabpa and Gabpb1 peaks. (J) Venn diagram showing intersect of GABP peaks (Fig. 3I), with the Gabpa and Gabpb1 regulon target genes (Fig. 3A).