Figure 1.
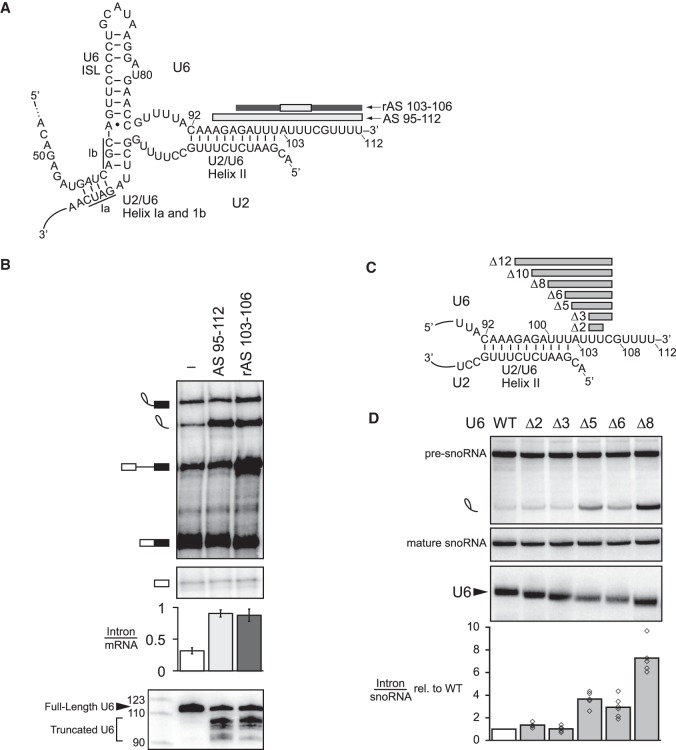
The 3′ end of U6 is required for turnover of excised, lariat intron. (A) Secondary structure of U6 and U2/U6 base-pairing interactions at the catalytic core of the spliceosome. Antisense oligos used for RNase H cleavage are depicted by bars above their target U6 sequences; light gray indicates DNA nucleotides in the anti-sense oligo, whereas dark gray indicates 2′-O-methyl nucleotides. (B) Truncation of the 3′ end of U6 by ∼10 nt impedes turnover of the excised intron. The panel shows denaturing PAGE analysis of radiolabeled ACT1 pre-mRNA following in vitro splicing in yeast extracts (yJPS860) that were first subjected to RNase H cleavage to truncate the 3′ end of U6 snRNA, directed by oligonucleotides depicted in A. Quantitation of intron turnover for each reaction was calculated as the molar ratio of excised intron to mRNA and is shown below the gel and is represented as the mean ± one standard deviation for three independent replicates. Cleavage of U6 was monitored by northern blot (bottom panel) with a radioactive probe directed to nucleotides 28–54 of U6. (C,D) Deletions of 5–8 nt in the 3′ end of U6 impede turnover of the excised lariat intron in vivo. Deleted residues are indicated in C. Viable deletions were analyzed by northern in D for pre-U3A snoRNA, the excised lariat intron, mature U3A snoRNA, and U6. Quantitation of excised intron levels, relative to mature U3A, is shown below the northern, after normalization to the levels in wild type; the average and values are shown for at least two biological replicates. See also Supplemental Figure S1.