In this Review, Gaertner et al. discuss how pancreatic cells differentiated from human pluripotent stem cells (hPSCs) have advanced our understanding of human pancreas development and disease and how technological advances are now beginning to enable large-scale disease modeling and screening for new therapeutics.
Keywords: embryonic, pancreatic development, stem cell
Abstract
A comprehensive understanding of mechanisms that underlie the development and function of human cells requires human cell models. For the pancreatic lineage, protocols have been developed to differentiate human pluripotent stem cells (hPSCs) into pancreatic endocrine and exocrine cells through intermediates resembling in vivo development. In recent years, this differentiation system has been employed to decipher mechanisms of pancreatic development, congenital defects of the pancreas, as well as genetic forms of diabetes and exocrine diseases. In this review, we summarize recent insights gained from studies of pancreatic hPSC models. We discuss how genome-scale analyses of the differentiation system have helped elucidate roles of chromatin state, transcription factors, and noncoding RNAs in pancreatic development and how the analysis of cells with disease-relevant mutations has provided insight into the molecular underpinnings of genetically determined diseases of the pancreas.
The pancreas encompasses both endocrine and exocrine compartments with important functions for nutrient metabolism. The endocrine pancreas is comprised of five cell types clustered in the pancreatic islets that regulate blood glucose homeostasis: glucagon-producing alpha cells, insulin-producing beta cells, somatostatin-producing delta cells, pancreatic polypeptide-producing gamma cells, and ghrelin-producing epsilon cells. The exocrine pancreas consists of acinar and ductal cells, which secrete the digestive enzyme-containing pancreatic juice and transport it into the duodenum, respectively. Several chronic diseases are associated with dysfunction of pancreatic cells, the most notable being diabetes mellitus, in which beta cells are destroyed by the immune system (type 1 diabetes; T1D) or functionally impaired (type 2 diabetes; T2D). Our current understanding of disease mechanisms has mostly been gained from the analysis of mouse models. While the molecular pathways that govern pancreas development are largely conserved between rodents and humans, there are also important differences (for detailed review, see Jennings et al. 2015; Nair and Hebrok 2015). For example, the first endocrine cells to arise in human development are beta cells, whereas alpha cells are the first to develop in mice. As discussed in detail in this review, a number of mutations that are associated with human diabetes do not cause diabetes in mice. Therefore, human cell models are needed to understand disease mechanisms and to develop novel therapies. Over the past 15 years, protocols have been developed to generate the different cell types of the pancreas from human pluripotent stem cells (hPSCs). In this review, we discuss how pancreatic cells differentiated from hPSCs have advanced our understanding of human pancreas development and disease and how technological advances are now beginning to enable large-scale disease modeling and screening for new therapeutics.
Recapitulating human pancreas development in vitro
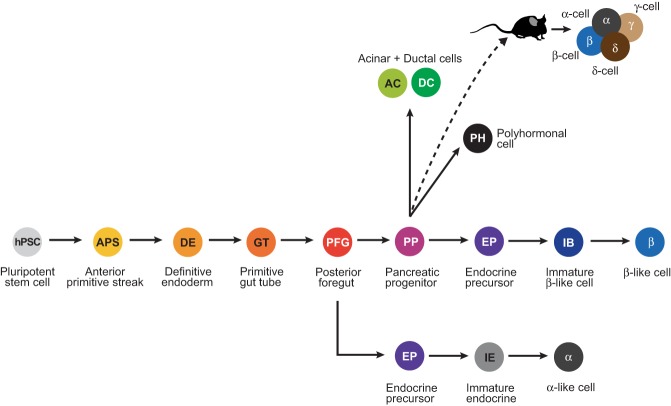
Compared to the detailed knowledge that exists in mice, the molecular mechanisms of human pancreas development are still largely unknown. This is especially true for early developmental stages that include induction of the endoderm germ layer, formation and patterning of the gut tube, and pancreas organ formation. The limiting factors for studying human pancreas development are the limited availability of embryonic and fetal tissue samples and, until the advent of single cell technologies, the cellular heterogeneity found within these tissues. The development of protocols for the differentiation of hPSCs towards the pancreatic lineage has provided a readily available and scalable human model in which to explore the basis of human development (Fig. 1).
Figure 1.
In vitro differentiation of human pluripotent stem cells (hPSCs) into different cell types of the pancreas. Overview of the stages of in vitro differentiation into the different pancreatic cell types, namely alpha, beta, acinar, and ductal cells. Beta cell differentiation protocols produce some monohormonal insulin+ cells that are glucose-responsive (beta-like cells) as well as nonfunctional insulin+ cells coexpressing other pancreatic hormones (polyhormonal cells). Upon implantation into mice, hPSC-derived pancreatic progenitors differentiate into glucose-responsive, insulin-secreting beta cells, as well as other endocrine cell types.
The current framework for our understanding of pancreas development has been mostly derived from studies in vertebrate model organisms. This knowledge has served as the guide to devise protocols for the stepwise differentiation of hPSCs into pancreatic progenitor cells (D'Amour et al. 2006; Kroon et al. 2008). For example, the use of Wnt agonists in combination with the TGFβ receptor ligand activin A to induce definitive endoderm from hPSCs is based on the finding in vertebrate embryos that both Wnt and Tgfβ signaling are required for definitive endoderm formation (Liu et al. 1999; Brennan et al. 2001). Likewise, observations in mice that pancreas induction requires inhibition of sonic hedgehog (Shh) (Hebrok et al. 2000) and that bone morphogenic protein (BMP) signaling favors the hepatic at the expense of the pancreatic fate (Rossi et al. 2001) have led to the use of Shh and BMP inhibitors for in vitro pancreas induction.
Despite significant advances in recapitulating pancreatic development in vitro, there are still shortcomings, particularly in regard to the generation of mature pancreatic cell types. The potential of having an unlimited cell source for diabetes cell replacement therapy has driven most efforts into generating insulin-producing beta cells from hPSCs. With current protocols, it is now possible to differentiate hPSC-derived pancreatic progenitors into insulin-producing cells that express key beta cell transcription factors, such has PDX1, NKX6.1, and MAFA (Pagliuca et al. 2014; Rezania et al. 2014; Russ et al. 2015). While beta-like cells generated with these protocols resemble human cadaveric beta cells in many aspects, up until recently they still lacked hallmark functional features of mature beta cells, which is glucose-stimulated insulin release and potentiation of insulin release by the gut-derived hormone GLP-1 (Rezania et al. 2014). Recent refinements of the protocol, most notably the omission of TGFβ signaling inhibitors at the beta cell maturation stage, now permit the generation of glucose-responsive beta cells in vitro (Nair et al. 2019; Velazco-Cruz et al. 2019; Veres et al. 2019). It is important to note that these refined protocols were optimized to predominately produce beta cells for use in cell therapy and that other pancreatic cell types are not generated to the same extent as seen during embryonic development. This could limit the utility of these protocols for studying developmental mechanisms. Separate protocols for generating alpha cells from hPSCs have also been developed (Rezania et al. 2011). These cells secrete glucagon in the presence of low glucose concentrations, whereas under high glucose conditions, secretion is decreased, suggesting physiological regulation of glucagon release. Given the contribution of dysregulated glucagon secretion to hyperglycemia in diabetes (Gromada et al. 2018), these cells could be a useful tool for identifying compounds that decrease glucagon secretion.
Recently, increasing efforts have also been put toward developing protocols to generate cells of the pancreatic exocrine lineage. The first reiteration of such a protocol generated organoids containing 0.5%–1% acinar cells and 10%–15% ductal cells, which the authors used to model pancreatic cancer initiation in vitro (Huang et al. 2015). Kleger and colleagues subsequently developed an improved protocol generating at least 19% acinar cells and 42% ductal cells that showed activity of the exocrine enzymes carbonic anhydrase, amylase, trypsin, and elastase (Hohwieler et al. 2017). Another ductal cell differentiation protocol was developed by Chen and colleagues to study ductal cell defects in patients with cystic fibrosis using patient-derived human induced pluripotent stem cells (iPSCs) (Simsek et al. 2016). Combined, these directed differentiation protocols have provided the field with human models to study the development and function of pancreatic endocrine and exocrine cells as well as to identify disease mechanisms.
Gene regulatory mechanisms of human pancreas development
Insights from transcriptional profiling
Transcriptional profiling of hPSCs during the stepwise in vitro differentiation toward the pancreatic endocrine cell fate has provided insight into how closely in vitro differentiation resembles in vivo processes. Such analysis has also led to the discovery of novel genes and pathways that govern human pancreas development. Initial analyses employed RNA-seq profiling of entire cell populations at different stages of differentiation (Xie et al. 2013; Loh et al. 2014; Cebola et al. 2015; Jiang et al. 2015). These studies revealed dynamic expression of regulators of endoderm and pancreas development in a pattern consistent with prior knowledge from studies in model organisms (Shih et al. 2013). The transcriptional profiles provided the first comprehensive atlas of stage-specific gene expression signatures during human endoderm development and identified novel candidate lineage-determining transcription factors, epigenetic regulators, and noncoding RNA species. Comparison of transcriptomes from hPSC cultures at the pancreatic progenitor cell stage to transcriptomes from microdissected pancreatic buds from human embryos further revealed overall high correlation of gene expression profiles (Cebola et al. 2015; Jennings et al. 2017). This suggested close similarity of the in vitro differentiated cells to their in vivo counterparts, at least until the pancreatic progenitor cell stage. In vitro-derived pancreatic progenitors more closely resembled microdissected dorsal than ventral pancreatic buds, suggesting induction of a dorsal rather than a ventral pancreatic program in vitro (Jennings et al. 2017). One limitation of these comparisons is the cell heterogeneity within dissected pancreatic buds (Ramond et al. 2017, 2018), which precludes identification of small, but possibly functionally meaningful, differences in gene expression between in vitro-generated cells and progenitors in the human embryo.
To better define cell populations arising during in vitro differentiation toward pancreas and beta cells, several groups employed transcriptome analyses at the single cell level (Petersen et al. 2017; Krentz et al. 2018; Sharon et al. 2019b; Veres et al. 2019). These analyses revealed fairly uniform cell populations throughout differentiation to the pancreatic progenitor cell stage (Sharon et al. 2019b). However, with the induction of endocrine cell differentiation, the cultures become highly heterogeneous, comprising different endocrine cell populations, some exocrine cells, as well as undifferentiated progenitors. This heterogeneity was observed across different beta cell differentiation protocols and hPSC lines. In late endocrine cell stage cultures, only a very small population of insulin-expressing cells exhibited gene expression profiles with high similarity to primary human beta cells (Petersen et al. 2017), showing substantial heterogeneity within the beta cell population. Whether refined protocols that promote the acquisition of mature functional features in hPSC-beta cells during prolonged culture (Velazco-Cruz et al. 2019; Veres et al. 2019) diminish this heterogeneity remains to be studied. A major challenge is to determine the extent to which these cell populations represent bona fide developmental intermediates in the human embryo. A population of cells observed across all studies coexpressed transcripts for insulin and glucagon (Petersen et al. 2017; Krentz et al. 2018; Sharon et al. 2019b; Veres et al. 2019). This polyhormonal population decreased over time of in vitro differentiation. Single cell gene expression analysis revealed a similar population in the human embryonic pancreas (Ramond et al. 2018); still, whether polyhormonal cells represent a normal developmental intermediate during pancreatic endocrine cell development in humans requires further study.
To gain insight into the developmental trajectory of endocrine progenitors in vitro, groups have either studied transcriptional profiles of these progenitors using hPSC lines that express GFP under control of regulatory sequences for the endocrine progenitor cell marker gene NEUROG3 (Petersen et al. 2017; Krentz et al. 2018) or reconstructed the lineage trajectory in silico from single-cell transcriptomic data (Sharon et al. 2019b; Veres et al. 2019). These studies revealed two waves of endocrine cell differentiation in vitro: early endocrine progenitors predominately adopted alpha cell identity, whereas later arising progenitors were beta cell-biased. Cell transplantation experiments in mice corroborated these findings (Sharon et al. 2019a). A similar time-dependent lineage bias of pancreatic endocrine progenitors has been observed in mice (Johansson et al. 2007), indicating similarity of the in vitro system to in vivo biology. However, the analysis of tissue sections from human fetal pancreas has shown that the pattern of NEUROG3 expression differs substantially between mice and humans (Jennings et al. 2013). Therefore, it remains unclear whether temporally separated waves of endocrine progenitors biased towards different lineages also exist during human pancreas development.
Single cell transcriptomic profiling of cells during the differentiation time course has provided novel insight into the mechanisms of human endocrine cell development. Sneddon and colleagues (Byrnes et al. 2018) identified a previously undescribed cell population marked by the transcription factor FEV as a lineage intermediate between the endocrine progenitor and endocrine cell states. This cell population has been consistently observed in early endocrine stage cultures using different hPSC lines and protocols (Petersen et al. 2017; Krentz et al. 2018; Veres et al. 2019). Additional studies of pancreata from mouse and human embryos further confirmed the existence of this cell population (Byrnes et al. 2018; Ramond et al. 2018; Bastidas-Ponce et al. 2019). This example illustrates that current in vitro differentiation protocols recapitulate significant in vivo events and can reveal new regulators of human pancreatic development. The identification of a role for the Wnt and Hippo signaling pathways in endocrine cell differentiation further demonstrates conserved features of the hPSC differentiation system (Cebola et al. 2015; Rosado-Olivieri et al. 2019; Sharon et al. 2019b). However, caution must be exercised, as differences from human development clearly exist. For example, unlike their in vivo counterparts (Ramond et al. 2018), hPSC-derived pancreatic progenitors express the foregut marker CDX2 (Petersen et al. 2017; Veres et al. 2019). Likewise, a population resembling enterochromaffin cells of the gut has been described in endocrine stage in vitro cultures (Veres et al. 2019). Whether this population exists during human pancreatic development is unknown. At present, we do not fully understand to what extent the in vivo lineage trajectory of pancreatic cells is conserved in the in vitro system. A key limitation in this regard is the lack of information about the lineage trajectory of pancreatic cells during human development. Insights gained from integrative analysis of single cell RNA-seq data from mouse embryonic pancreas illustrate how such data can be used to reconstruct cell lineage relationships (Byrnes et al. 2018; Scavuzzo et al. 2018). Similar single cell-resolved time course data throughout human pancreas development would be a tremendous resource for benchmarking cells derived during in vitro differentiation.
Noncoding RNAs
With advances in high-throughput sequencing technologies came the realization that a much larger than anticipated fraction of mammalian genomes is transcribed into RNA. While only about 2% of genomic DNA gives rise to all protein-coding RNAs, a much larger portion of the genome is pervasively transcribed into diverse and loosely defined classes of noncoding RNAs (ncRNAs) whose functional significance is largely unclear (Carninci et al. 2005; Palazzo and Lee 2018). Among ncRNAs, microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) constitute the most extensively studied groups. miRNAs encompass ∼22 nucleotides (nt), whereas lncRNAs are defined by a length >200 nt. miRNAs regulate mRNA stability and/or translation through the base-pairing of their seed sequences with binding sites in the 3′-untranslated regions (3′-UTRs) of their target mRNAs, thus posttranscriptionally fine-tuning expression of hundreds of mRNAs (Martinez-Sanchez et al. 2016; O'Brien et al. 2018). lncRNAs, on the other hand, are thought to function as molecular scaffolds that interact with both proteins and nucleic acids, which enables them to interface with any aspect of gene regulation (Esguerra and Eliasson 2014).
While there are a few examples of ncRNAs with roles in mature beta cells, controlling beta cell fate maintenance or regulating insulin secretion (Morán et al. 2012; Liang et al. 2015; Mi et al. 2015; Arnes et al. 2016; Akerman et al. 2017; Motterle et al. 2017), their role during endocrine cell differentiation is less understood. It is known that miRNAs play an important role in beta cell development, as conditional loss of the miRNA-processing ribonuclease Dicer1, which globally impairs miRNA biogenesis, in the embryonic mouse pancreas results in decreased beta cell mass (Lynn et al. 2007). However, little is known about the individual miRNAs that are involved. miR-26, miR-7, and miR-375 have been identified as regulators of beta cell differentiation (Kloosterman et al. 2007; Nieto et al. 2012; Fu et al. 2013), but their precise function is still unknown. Two groups have used hPSCs to functionally annotate miRNAs expressed during pancreatic endocrine cell differentiation (Chen et al. 2011; Liao et al. 2013), with the aim to computationally identify miRNA target mRNAs that regulate pancreas development. Both studies defined distinct sets of miRNAs whose expression is negatively correlated with their putative target mRNAs; however, the overlap between these sets is very low, presumably owing to differences in the differentiation protocols and computational methods used to identify target mRNAs. Laurent and colleagues (Liao et al. 2013) performed luciferase reporter assays to validate these predicted miRNA-mRNA interactions and their roles in regulating differentiation. They found that expression of the pancreatic progenitor-expressed transcription factor RFX6, known to regulate pancreatic endocrine cell development (Smith et al. 2010), is repressed by miR-30d and let-7e, two miRNAs induced during endocrine cell differentiation. Recently, our group combined small RNA sequencing, miRNA overexpression in hPSC-derived pancreatic progenitors, and network modeling approaches to characterize miRNA regulatory networks active in human endocrine cell differentiation. The analysis identified let-7g, let-7a, miR-200a, and miR-375 as endocrine-enriched miRNAs that target cell cycle-associated transcription factors and act to promote cell cycle exit and beta cell differentiation (Jin et al. 2019). Consistent with prior observations that miRNAs fine-tune gene expression rather than acting as genetic switches (Vidigal and Ventura 2015), the effects of overexpressing endocrine cell-enriched miRNAs on gene expression and endocrine cell differentiation were relatively small.
The role of lncRNAs in human pancreas development is even less explored than the role of miRNAs. In contrast to miRNAs, lncRNAs have low sequence conservation between species. Moreover, lncRNAs tend to be expressed in a highly cell type-specific manner and may have context-dependent functions, properties that necessitate functional studies in relevant human cell types. To date, most studies of lncRNAs during human endoderm development have focused on definitive endoderm, and the role of lncRNAs in pancreas development remains to be defined. An emerging conclusion from all studies of lncRNAs in developmental contexts is that lncRNAs regulate the expression of their neighboring genes, which commonly encode lineage-determining transcription factors (Sigova et al. 2013; Herriges et al. 2014; Daneshvar et al. 2016; Luo et al. 2016). Targeted lncRNA silencing in hPSCs combined with transcriptomic analysis identified DEANR1 as a lncRNA critical for endoderm differentiation by activating expression of the nearby gene encoding the transcription factor FOXA2. DEANR1 regulates FOXA2 expression by recruiting SMAD2/3 to the FOXA2 promoter (Jiang et al. 2015). Similarly, the lncRNA DIGIT modulates definitive endoderm formation by inducing the expression of Goosecoid, a homeobox transcription factor known to regulate endoderm development (Daneshvar et al. 2016). Additional work is needed to catalog the lncRNAs expressed during human pancreatic development and to define their roles in pancreatic cell differentiation. In the future, the principles learned from such studies could aid the development of direct reprogramming strategies, e.g., by deploying ncRNAs as reprogramming factors that help establish beta cell-specific gene expression programs in developmentally related cell types.
The role of chromatin state in pancreatic differentiation
The relative uniformity of cells generated during the stepwise differentiation of hPSCs into pancreatic progenitors (Sharon et al. 2019b) provides a model system to map chromatin dynamics during the progression of hPSCs through definitive endoderm toward pancreas. Furthermore, given the defined growth factor composition of the culture medium at every step of differentiation, the system provides a platform to study how chromatin state affects sequential signal-dependent gene expression changes during developmental progression. Chromatin state controls access of regulatory proteins to DNA and shapes the three-dimensional organization of the genome, e.g., by supporting or restricting long-range contacts between enhancers and their target gene promoters (Venkatesh and Workman 2015). Covalent posttranslational modifications of histone proteins can affect chromatin structure either directly or by recruiting effector proteins that recognize specific modifications (Fig. 2). During development, gene loci of important developmental regulators often exhibit changes in chromatin state before gene transcription is initiated. Chromatin priming is thought to be an important mechanism that facilitates correct temporal regulation of gene expression in response to cell extrinsic signals. At the level of promoters, co-occurrence of the activating histone H3 lysine 4 trimethylation (H3K4me3) and repressive histone H3 lysine 27 trimethylation (H3K27me3) specifies bivalent poised chromatin (Bernstein et al. 2006). In hPSCs, bivalent chromatin is a hallmark of developmental control genes, poising these genes for rapid activation during differentiation correlated with removal of H3K27me3. Chromatin priming also occurs at the level of transcriptional enhancers, which regulate transcription of their target genes at a distance and play a key role in cell type-specific gene regulation (Catarino and Stark 2018). Enhancers can be identified genome-wide based on the presence of histone H3 lysine 4 monomethylation (H3K4me1) and histone H3 lysine 27 acetylation (H3K27ac). The presence of H3K4me1 in the absence of H3K27ac marks inactive but poised enhancers, whereas in combination with H3K27ac, it signifies active enhancers. Studies have shown that acquisition of a poised enhancer state during development is accompanied by a gain in chromatin accessibility (Stergachis et al. 2013; Lee et al. 2019), which signifies that recruitment of transcription factors precedes enhancer activation. The analysis of sequence motifs enriched at enhancers can help identify the transcription factors involved in enhancer regulation.
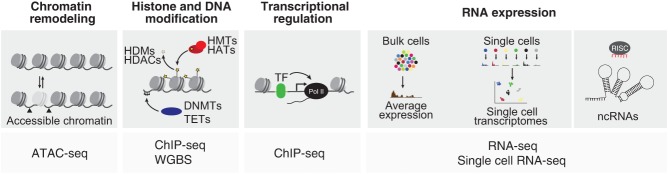
Figure 2.
Genome-scale next generation sequencing-based assays employed to study gene regulatory mechanisms of human pancreas differentiation. Schematic showing different genome-scale assays used to analyze chromatin remodeling, histone and DNA modifications, transcriptional regulation, gene expression, and expression of noncoding RNAs during pancreatic differentiation. (DNMTs) DNA methyltransferases, (TET) ten-eleven translocation enzymes, (HMTs) histone methlytransferases, (HATs) histone acetyltranferases, (TF) transcription factor, (RISC) RNA-induced silencing complex, (ATAC-seq) assay for transposase-accessible chromatin using sequencing, (WGBS) whole genome bisulfide sequencing, (ChIP-seq) chromatin immunoprecipitation sequencing.
Several studies have systematically mapped dynamics in histone modifications, chromatin accessibility, and DNA (hydroxy)methylation during hPSC differentiation into pancreatic progenitors (Xie et al. 2013; Loh et al. 2014; Wang et al. 2015; Li et al. 2018). Analysis of bivalent promoters revealed a bivalent state at master regulators of definitive endoderm and pancreas formation in hPSCs (Xie et al. 2013). These promoters lost the repressive H3K27me3 mark in a stage-specific manner, concomitant with initiation of gene transcription at the associated target gene. Thus, a hallmark feature of lineage-determining transcription factors during endodermal development is their bivalent state in hPSCs and stage-specific activation through removal of H3K27me3. The analysis not only validated this pattern for virtually all known regulators of pancreas development, but also revealed new candidates, such as OVOL1 and MEIS1, whose function in pancreas development remains to be studied. Important pancreatic transcription factors exhibited a similar pattern of H3K27me3 dynamics during murine endoderm development (Xu et al. 2014). However, the analysis of H3K27me3 in murine progenitors also revealed differences from hPSC-derived pancreatic progenitors, particularly at genes involved in organ morphogenesis. Given that morphogenetic processes associated with pancreatic organ development are not recapitulated by current hPSC differentiation protocols, this finding is not surprising and points to a limitation of the in vitro system for studying all aspects of in vivo development. Accordingly, direct comparison of transcriptional profiles from hPSC-derived pancreatic progenitors and human fetal pancreatic buds showed differences in the expression of genes associated with morphogenetic processes (Jennings et al. 2017).
The analysis of chromatin dynamics throughout the differentiation time course also revealed chromatin priming at the level of enhancers. At the gut tube stage, enhancers specific to gut tube-derived organ lineages (i.e., lung, pancreas, and liver) exhibited en masse de novo deposition of H3K4me1 (Wang et al. 2015), signifying poised chromatin. Experimentally, the acquisition of this poised enhancer state predicted the ability of endodermal intermediates to respond to inductive signals, suggesting a role for chromatin priming at enhancers in the acquisition of developmental competence. In addition to H3K4me1 deposition, other mechanisms of chromatin priming at developmental enhancers may exist. In hPSCs, Lim and colleagues (Loh et al. 2014) identified presence of the histone variant H2AZ at enhancers that become active in definitive endoderm. H2AZ may facilitate nucleosome displacement and enhancer activation (Brunelle et al. 2015) by destabilizing histone octamers (Jin and Felsenfeld 2007). Given the importance of enhancers in developmental gene regulation, it comes as no surprise that mutations in enhancers can cause developmental defects and disease. The intersection of enhancer annotations from hPSC-derived pancreatic progenitors with whole-genome sequences from individuals with pancreatic agenesis revealed mutations in a hitherto uncharacterized pancreatic progenitor-specific enhancer for the pancreatic transcription factor PTF1A in several families (Weedon et al. 2014). This example illustrates how integration of genome sequencing and epigenomic annotation in hPSC-derived developmental intermediates can help identify mechanisms of human development and disease.
New insights into transcription factors governing pancreatic development
Enhancers regulate gene transcription through the recruitment of transcriptional complexes comprised of DNA-binding transcription factors, coregulators, and components of the general transcriptional machinery (Spitz and Furlong 2012). The analysis of sequence motifs enriched at enhancers active in hPSC-derived pancreatic progenitors has provided insight into the transcription factors involved in the regulation of pancreatic development in humans (Cebola et al. 2015; Wang et al. 2015). Expectedly, human pancreatic progenitor-active enhancers were enriched for motifs of transcription factors identified as regulators of early pancreas development in model organisms, including PDX1 (Offield et al. 1996), GATA transcription factors (Carrasco et al. 2012; Xuan et al. 2012), SOX9 (Seymour et al. 2007), and HNF1B (Haumaitre et al. 2005). ChIP-seq analysis in hPSC-derived pancreatic progenitors further demonstrated that these transcription factors reside jointly in transcriptional complexes at enhancers, providing a mechanism for their cooperative roles in specification of the pancreatic lineage (Shih et al. 2015). The analysis of sequence motifs enriched at pancreatic progenitor enhancers also revealed novel regulators of pancreatic development. Ferrer and colleagues identified the transcription factor TEAD as a component of the transcriptional complexes at early pancreatic enhancers (Cebola et al. 2015). TEAD is a downstream effector of Hippo signaling and acts together with its coactivator YAP. In pancreatic progenitors, YAP activity is regulated by cues from the extracellular matrix and couples extrinsic mechano-signals to the regulation of gene transcription (Mamidi et al. 2018). Functional studies of the YAP-TEAD complex in hPSCs during pancreatic differentiation demonstrated an important role for YAP-TEAD in the regulation of pancreatic progenitor cell expansion and endocrine cell differentiation (Cebola et al. 2015; Mamidi et al. 2018; Rosado-Olivieri et al. 2019). The identification of TEAD and YAP as components of the transcription factor complexes at early pancreatic enhancers illustrates how cues from the extracellular environment are integrated with lineage identity cues at the level of enhancers.
The observation that pancreatic enhancers acquire a poised chromatin state in gut tube intermediates prior to enhancer activation (Wang et al. 2015) raised the question of which transcription factors could mediate enhancer priming during endoderm development. Motif analysis followed by ChIP-seq experiments identified FOXA1 and FOXA2 as enriched at pancreatic enhancers in gut tube intermediates (Wang et al. 2015). FOXA transcription factors belong to the class of so-called “pioneer” transcription factors, which can recognize their target sites in the context of nucleosomal DNA and closed chromatin (Zaret and Carroll 2011). The ability of FOXA transcription factors to actively displace histones in local chromatin, thus conferring competence for other factors to bind, suggests that FOXA1 and FOXA2 could mediate chromatin priming of pancreatic enhancers during endoderm development. Confirming this prediction, a recent study showed that FOXA2 deletion in hPSCs indeed prevents early chromatin priming of pancreatic enhancers and consequently pancreatic lineage induction (Lee et al. 2019). Given that FOXA1/2 recruitment coincides with the acquisition of responsiveness to lineage-inductive signals during pancreas differentiation, it is likely that FOXA-bound enhancers are also signal responsive. It remains to be studied how lineage-inductive cues selectively activate organ-specific enhancers for lung, pancreas, and liver in the context of a broadly primed chromatin landscape at enhancers for all organ lineage derived from the gut tube (Wang et al. 2015). It is possible that organ-specific activation of primed enhancers is achieved as a result of differences in enhancer sequence architecture between organs. This could explain why pancreatic enhancers are activated in response to Shh inhibition and retinoic acid, whereas liver enhancers are activated in response to BMP and FGF. There is evidence for such a mechanism in definitive endoderm development, where occupancy of the endoderm transcription factors GATA6 and EOMES has been shown to coincide with recruitment of SMAD2/3, which translocates to the nucleus in response to endoderm induction by activin A (Chia et al. 2019).
If developmental competence for all gut tube-derived organs is broadly established at the gut tube stage, the question arises of which mechanisms prevent the activation of genes associated with alternative fates once progenitors have committed to a specific lineage. Analysis of PDX1 binding sites in hPSC-derived pancreatic progenitors suggests that PDX1 is not only required for the activation of pancreatic genes, but also acts as a transcriptional repressor of genes linked to the liver fate (Teo et al. 2015), an observation that is consistent with previously noted ectopic expression of hepatic genes in the embryonic pancreas of mice lacking Pdx1 or Sox9 (Seymour et al. 2012). Further supporting the idea that lineage-determining transcription factors suppress inducers of alternative fates, PDX1 and SOX9 have been shown to occupy regulatory sequences of intestinal lineage determinants in hPSC-derived pancreatic progenitors (Shih et al. 2015). Thus, PDX1 and SOX9 may cooperatively both activate pancreatic genes and repress intestinal and liver genes. In the future, the hPSC differentiation system could be employed to globally identify genes involved in individual steps of differentiation, using genome-scale CRISPR screens. Recently, one such screen in hPSCs has identified the JNK–JUN pathway as a barrier for definitive endoderm formation (Li et al. 2019). Inhibition of JNK improved the efficiency of definitive endoderm induction protocols and increased the percentage of cells committing to the pancreatic and lung lineages. This example illustrates how knowledge from genome-wide screens can help improve existing differentiation protocols.
Pancreatic disease modeling with hPSCs
Most of our current knowledge about how disease-associated genes cause specific phenotypes has been derived from studies in model organisms. However, commonly utilized rodent models in many cases fail to recapitulate the disease phenotypes observed in humans (Teo et al. 2016; Zhu et al. 2016; Shi et al. 2017; Tiyaboonchai et al. 2017). A major limitation for the use of primary human cells to model disease is their limited availability and variable quality after isolation, as is the case for human cadaveric beta cells. Moreover, terminally differentiated cells do not permit examination of phenotypes arising during development. With respect to both of these challenges, hPSC-derived cells pose an attractive model for dissecting genetic disease mechanisms.
Modeling monogenic forms of diabetes
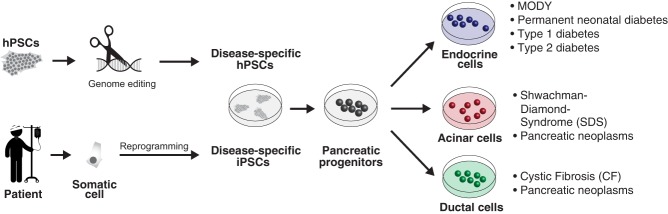
Relevant cell types for modeling human disease can be generated from hPSCs via different methods (Fig. 3). First, hPSCs can be directly isolated from early human embryos following pre-implantation genetic diagnosis (Ben-Yosef et al. 2008; Stephenson et al. 2009; Tropel et al. 2010). A second, more commonly used method is to reprogram fibroblasts from affected individuals into iPSCs. Here, correction of the mutation by gene editing techniques is an important control to demonstrate causality between the mutation and the observed phenotype. Finally, disease-relevant mutations can be introduced directly into existing hPSC lines via genome editing (Maury et al. 2012). Genetic diseases related to all pancreatic lineages (endocrine, acinar, and ductal) have been modeled using hPSCs. Due to their central role in the pathogenesis of diabetes, the largest effort has been in modeling diseases affecting pancreatic beta cells.
Figure 3.
Approaches to model genetic causes of pancreatic diseases. Disease-associated mutations or variants are introduced into hPSCs via genome editing, or patient-specific iPSCs are generated via reprogramming. These approaches have been employed to model endocrine and exocrine diseases. (hPSCs) human pluripotent stem cells, (iPSCs) induced pluripotent stem cells, (MODY) maturity onset diabetes of the young.
Biallelic mutations of many pancreatic lineage-determining transcription factors, including PDX1, PTF1A, NGN3 (encoded by NEUROG3), GLIS3, RFX6, NEUROD1, MNX1, and NKX2.2 cause permanent neonatal diabetes in humans (Stoffers et al. 1997; Sellick et al. 2004; Senée et al. 2006; Rubio-Cabezas et al. 2010, 2011; Smith et al. 2010; Dimitri et al. 2011; Bonnefond et al. 2013; Flanagan et al. 2014a,b). The functions of these transcription factors have been extensively studied in mice (Romer and Sussel 2015), but their specific roles in human pancreas development had remained elusive. Huangfu and colleagues employed genome editing in hPSCs to systematically characterize loss-of-function phenotypes of eight pancreatic transcription factors during in vitro pancreatic differentiation (Zhu et al. 2016). The study demonstrated conserved requirements for HES1, ARX, RFX6, PDX1, and NGN3 in pancreatic development and cell differentiation between mice and humans. Interestingly, the study also identified potentially divergent roles of some transcription factors in mice and humans. For example, biallelic frameshift mutations in PTF1A and MNX1, transcription factors necessary for pancreatic organ formation in mice (Harrison et al. 1999; Li et al. 1999; Fukuda et al. 2008), surprisingly did not affect endocrine cell differentiation of hPSCs. Similarly, hPSCs carrying a homozygous loss-of-function mutation in GLIS3 exhibited no endocrine differentiation defects, contrary to Glis3 null mutant mice, which have reduced endocrine cell numbers (Kang et al. 2009). It is important to note that the observed phenotypes in mutant hPSCs were not always consistent across studies from different laboratories. For example, in contrast to Zhu's study (Zhu et al. 2016), Chen and colleagues reported reduced numbers of insulin+ cells and increased cell death in GLIS3−/− hPSC cultures (Amin et al. 2018). Similarly, NGN3−/− hPSCs produced some insulin+ cells upon in vitro differentiation in one study (Zhu et al. 2016), whereas another study reported a complete endocrine cell differentiation block in NGN3−/− hPSCs (McGrath et al. 2015). The underlying cause for these discrepancies is still unclear. Possible explanations include divergent differentiation protocols between laboratories and differences in the genetic background of hPSC lines. The potential protocol-dependency of phenotypes illustrates limitations of in vitro hPSC differentiation systems for modeling the impact of genetic mutations on human development.
Nevertheless, the analysis of transcription factor-deficient hPSCs during in vitro differentiation has helped define mechanisms whereby mutations in pancreatic transcription factors could cause pancreatic abnormalities and diabetes. Analysis of RFX6−/− cells revealed a previously unknown early developmental role for RFX6 in the formation of PDX1+ pancreatic progenitor cells (Zhu et al. 2016), which helps explain why homozygous RFX6 mutations in humans are associated with pancreas hypoplasia (Concepcion et al. 2014; Zegre Amorim et al. 2015). Furthermore, it was shown that monoallelic PDX1 mutations cause a reduction in insulin+ cells (Zhu et al. 2016), suggesting a dosage-sensitive requirement for PDX1 in human pancreas development that may predispose affected carriers to developing diabetes later in life (Staffers et al. 1997). Corroborating these findings, Lickert and colleagues modeled heterozygous mutations in the PDX1 transactivation domain found in patients with increased risk for diabetes and observed impaired beta cell development and function in hPSC models (Wang et al. 2019). Similarly, hPSC models helped define the mechanism whereby activating mutations in the transcription factor STAT3 cause a form of permanent neonatal diabetes that is characterized by both pancreatic hypoplasia and beta cell autoimmunity (Otonkoski et al. 2000; Flanagan et al. 2014b). In vitro modeling with patient iPSCs demonstrated an immune-independent developmental defect caused by precocious upregulation of NEUROG3 that leads to premature differentiation of alpha cells at the expense of beta cells and depletion of pancreatic progenitor cells (Saarimäki-Vire et al. 2017).
Several monogenic forms of diabetes in humans are not recapitulated in mouse models. One such example is monoallelic loss-of-function mutations in the transcription factor GATA6, which in humans lead to pancreas agenesis and consequently neonatal diabetes (Allen et al. 2014), but in mice cause no pancreatic defect (Carrasco et al. 2012; Xuan et al. 2012). To gain insight into the role of GATA6 in human pancreas development, three independent groups studied effects of GATA6 mutations in an in vitro hPSC model of pancreatic cell differentiation (Shi et al. 2017; Tiyaboonchai et al. 2017; Chia et al. 2019). All three studies found that homozygous GATA6 loss-of-function mutations caused a defect in definitive endoderm formation that would not be compatible with life. The effects of heterozygous GATA6 mutations on definitive endoderm development were less consistent across studies. While Vallier and colleagues noted an ∼25% decrease in cells adopting the definitive endoderm fate, the groups of Danwei Huangfu and Paul Gadue observed no difference between GATA6+/− and control cells at the definitive endoderm stage. This difference is unlikely to be due to different mutations being analyzed, as the studies by Vallier and Huangfu both analyzed mutations that did not produce a truncated protein. All three studies found that heterozygous mutations in GATA6 impaired the formation of endocrine-specified pancreatic progenitors leading to reduced numbers of insulin+ cells in late stage cultures. Thus, these studies clearly demonstrate a requirement for two functional copies of GATA6 for normal endocrine cell development in humans. Interestingly, Gadue and colleagues showed that the degree to which beta cell differentiation is affected by loss of GATA6 can be modulated by varying the concentration of pancreas differentiation factors in the culture medium (Tiyaboonchai et al. 2017). This illustrates that phenotypes depend, at least to some extent, on the media composition used during in vitro differentiation. Protocol differences might also explain why one study found GATA6 haploinsufficiency to impair glucose-stimulated insulin secretion (Tiyaboonchai et al. 2017), while the other study did not (Shi et al. 2017).
Maturity onset diabetes of the young (MODY) is a monogenic autosomal dominant form of diabetes that manifests during childhood or early adulthood. As of today, at least 14 MODY genes have been described (Firdous et al. 2018), most of which encode transcription factors essential for beta cell development and function. In many cases it is unclear how MODY mutations cause insulin insufficiency, and mouse models have not always recapitulated the human phenotype. This is the case for MODY5, which is caused by mutations in the transcription factor HNF1B. While heterozygous loss-of-function mutations in HNF1B are associated with diabetes and pancreas hypoplasia in humans (Bellanné-Chantelot et al. 2004; Edghill et al. 2006; Haumaitre et al. 2006; Haldorsen et al. 2008), heterozygous Hnf1b mutant mice do not develop hypoplasia nor do they become diabetic (De Vas et al. 2015). In vitro differentiation of iPSCs from MODY5 patients revealed misexpression of transcription factors involved in beta cell development and reduced expression of the insulin gene activator PAX6, suggesting that loss of one copy of HNF1B in humans impairs beta cell development and function (Teo et al. 2016).
Similar to MODY5, the phenotypes of MODY3/HNF1A and MODY1/HNF4A mutations are not fully mirrored in mice (Stoffel and Duncan 1997; Pontoglio et al. 1998; Garcia-Gonzalez et al. 2016). Using genetically engineered hESCs, Gadue and colleagues showed that loss of one or two copies of HNF1A diverts endocrine differentiation from the beta cell towards the alpha cell fate (Cardenas-Diaz et al. 2019). Moreover, HNF1A-deficient beta cells were functionally compromised, exhibiting decreased oxidative phosphorylation and impaired insulin secretion. Teo and colleagues recently reported a defect in the development of foregut progenitors and impaired induction of insulin gene expression in iPSCs from MODY1/HNF4A patients (Ng et al. 2019). However, contrary to these findings, another study found beta cell development and insulin expression to be unaffected in a MODY1 iPSC model (Vethe et al. 2017). The reasons for the discrepancy are unclear, and additional studies are necessary to address how MODY1/HNF4A causes diabetes in humans.
Although most genes associated with monogenic diabetes encode transcription factors, other genes have also been identified. For example, common forms of permanent neonatal diabetes are linked to dominant negative insulin gene mutations that cause insulin protein misfolding in the endoplasmic reticulum (ER), inducing ER stress and eventually beta cell failure (Liu et al. 2010). To elucidate the pathogenic mechanisms of these mutations, patient iPSCs were differentiated into beta-like cells (Balboa et al. 2018). Single cell transcriptome analysis revealed increased expression of ER stress-associated genes and reduced expression of genes related to beta cell proliferation and function in in vitro-generated beta cells. Upon transplantation into mice, mutant beta cells exhibited signs of ER-stress as well as functional defects associated with dysregulated mTORC1 signaling, which is a critical pathway for beta cell maturation (Ni et al. 2017; Sinagoga et al. 2017). Elevated ER-stress is also the cause of beta cell failure in Wolfram syndrome, which is another form of monogenic diabetes. Wolfram syndrome is caused by mutations in the wolframin (WFS1) gene, which encodes an ER-resident protein that is highly expressed in beta cells. Although the molecular function of WFS1 is unknown, a recent study using patient iPSC-derived beta cells showed reduced insulin protein and secretory defects in mutant beta cells that were caused by activation of the ER unfolded protein response pathway (Shang et al. 2014). The findings suggest that WFS1 protects beta cells from ER stress to preserve their function. Together, these studies illustrate that hPSC models have begun to provide insights into disease mechanisms of human diabetes. Recent advances in generating glucose-responsive beta cells from hPSCs in vitro (Velazco-Cruz et al. 2019; Veres et al. 2019) will improve our ability to accurately model beta cell phenotypes in vitro and will likely yield additional insights into the mechanisms of beta cell failure in diabetes.
Modeling type 1 and type 2 diabetes
Most forms of diabetes are not caused by a single gene mutation but are polygenetic in etiology and result from a complex interplay between beta cells and their environment. T1D is characterized by autoimmune destruction of beta cells, resulting in decreased beta cell mass and insufficient insulin production. While the immune system plays a critical role in T1D, there is evidence that beta cell-intrinsic mechanisms also contribute to disease pathogenesis. Studies in the NOD mouse model of T1D have shown an immune-independent increase in the sensitivity of NOD beta cells to stress-induced cell death (Dooley et al. 2016). To study beta cells in T1D, several groups generated iPSCs from individuals with T1D and differentiated these cells into beta-like cells in vitro (Maehr et al. 2009; Thatava et al. 2013; Millman et al. 2016; Hosokawa et al. 2017; Manzar et al. 2017; Sui et al. 2018). As expected, beta-like cells could be readily derived from T1D-iPSCs, and these beta cells did not exhibit overt phenotypic defects. Furthermore, a small scale comparison of iPSC-beta-like cells from three T1D and three unaffected individuals showed no difference in cell death upon exposure to pro-inflammatory cytokines (Millman et al. 2016). In contrast, a study of beta-like cells derived from iPSCs generated from patients with fulminant T1D showed increased apoptosis following cytokine treatment (Hosokawa et al. 2017). Fulminant diabetes is a subclass of T1D characterized by rapid onset with less autoimmune contribution (Imagawa and Hanafusa 2006). While larger scale studies are clearly needed to draw conclusions, these studies illustrate that iPSCs could help provide mechanistic insight into the clinical heterogeneity of T1D. Moreover, an isogenic coculture model of iPSC-beta-cells and immune cells isolated from the same individual could prove informative for identifying mechanisms of beta cell destruction in T1D. When combined with drug screens, these models could pave the way for new therapeutic interventions.
Like T1D, T2D is polygenic in origin and its manifestation is strongly determined by environmental influences (Thomsen and Gloyn 2014). Genome-wide association studies have identified over 400 genomic risk loci for T2D (Mahajan et al. 2018). Most T2D risk signals are located in noncoding regions of the genome, which makes it difficult to identify the target genes affected by these risk variants. T2D risk-associated genetic variants are highly enriched in gene regulatory elements that are active in islets (Pasquali et al. 2014), suggesting a significant contribution of beta cell gene regulation to T2D pathogenesis. Since, compared to coding regions, regulatory sequences are less conserved between mouse and human, hPSC-derived beta cells could provide a model to study variant effects on gene regulation and to identify target genes of T2D risk-associated variants. One possible approach is to compare gene expression in human iPSC-beta cells from a large collection of iPSCs representing common risk variants and reference alleles. Such genetically well-characterized collections of iPSCs already exist, and proof-of-concept studies suggest that these collections are useful for validating variant effects in metabolic disease (Warren et al. 2017). As an alternative approach, risk and non-risk variants can be introduced into hPSCs by gene editing.
In T2D, chronic exposure of beta cells to high blood glucose and fatty acids (FAs) induces prolonged activation of the ER stress response that can lead to beta cell apoptosis (Biden et al. 2014). To gain insight into the function of genes with previously suggested roles in T2D, Chen and colleagues used hPSCs to study the T2D genetic risk-associated genes CDKAL1, KCNQ1, and KCNJ11 (Zeng et al. 2016). Beta cells derived from hPSCs with biallelic loss-of-function mutations exhibited impaired insulin secretion in response to secretagogues. Furthermore, consistent with described roles of CDKAL1 in the ER stress response (Brambillasca et al. 2012), CDKAL−/− beta-like cells displayed increased ER stress and apoptosis under high glucose and FAs (Zeng et al. 2016). A follow-up study further identified the metallothionein (MT) protein MT1E as a downstream effector of CDKAL1 in the regulation of ER stress and beta cell function (Guo et al. 2017). These studies illustrate how hPSC models can help define disease mechanisms leading to T2D.
Modeling diseases associated with acinar and ductal cells
Various diseases arise from the exocrine compartment of the pancreas and, although protocols to generate acinar and ductal cells from hPSCs have been developed, exocrine cells have been studied in much less detail than pancreatic endocrine cells. One example of an exocrine disease that has been modeled in vitro is Shwachman-Diamond syndrome (SDS), which is characterized by exocrine pancreatic insufficiency that causes malabsorption and malnutrition. SDS is caused by mutations in the gene encoding the Shwachman-Bodian-Diamond syndrome protein (SBDS) that is involved in ribosome biogenesis (Burwick et al. 2012; Gijsbers et al. 2018). In vitro exocrine differentiation of hPSCs with a knock-down of SBDS or SDS-iPSCs revealed disorganized ductal cells as well as a loss of acinar cells through increased apoptosis with SBDS loss or mutation (Tulpule et al. 2013). These phenotypes appeared to result from aberrant cytotoxic protease secretion, as they could be rescued by supplementing protease inhibitors.
Cystic fibrosis (CF) is another example of a genetic disease that affects the exocrine compartment. Specifically, mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel gene that is expressed in pancreatic ductal cells lead to pancreatic fibrosis and exocrine insufficiency. To gain insight into CF pathophysiology, several groups employed iPSC differentiation strategies to model CF defects in vitro (Takizawa-Shirasawa et al. 2013; Huang et al. 2015; Simsek et al. 2016; Hohwieler et al. 2017). Taking this approach, Kleger and colleagues also tested compounds known to improve CFTR function on CF patient-iPSC-derived pancreatic organoids and found that they could indeed partially rescue fluid secretion into the lumen of CF organoids (Hohwieler et al. 2017). Exemplified by a recent report, hPSC-derived ductal and acinar cells could also provide insight into events that initiate the formation of pancreatic cancer and its precursor lesions (Huang et al. 2015).
Future perspectives
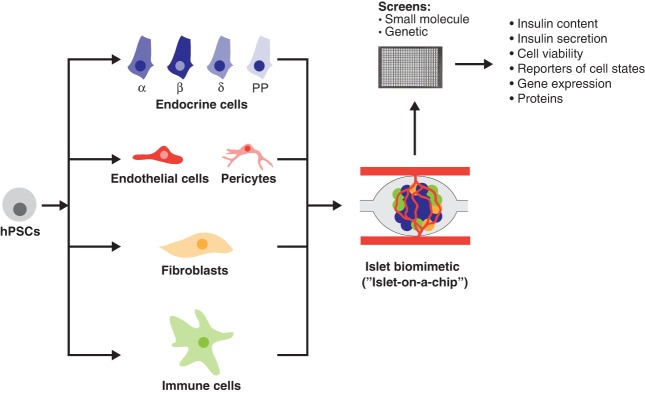
The most current beta cell differentiation protocols produce beta-like cells with functional features close to primary human beta cells (Velazco-Cruz et al. 2019; Veres et al. 2019). This recent leap forward now provides a nontransformed human beta cell model that can be adapted for high-throughput screens. The Chen laboratory has already conducted proof-of-concept studies that small molecule screens on hPSC-beta cells can help identify modifiers of disease processes in beta cells. A 2000 FDA-approved compound screen on CDKAL1−/− beta-like cells identified the c-Fos/AP-1 inhibitor T5224 as capable of reversing the cell death phenotype under high glucose and high FAs (Zeng et al. 2016). A similar screen on GLIS3−/− beta-like cells showed that the TGF-β inhibitor Galunisertib could prevent GLIS3 deficiency-induced beta cell death (Amin et al. 2018). hPSC-derived beta cells could not only help identify new drugs for the treatment of diabetes, but could also be employed to conduct CRISPR-Cas9-mediated screens to better characterize regulators of human beta cell health and to identify modifiers of disease processes (Fig. 4). To effectively use hPSC-beta cells for high-throughput screens, it will be important to further refine and scale existing platforms. Even with the most advanced protocols, there is still substantial cell heterogeneity in hPSC cultures. Enrichment strategies for mature beta cells, as recently described (Saunders et al. 2018; Velazco-Cruz et al. 2019), could help overcome this limitation. Finally, to appropriately model the rich cell–cell interactions that occur within the islet and contribute to diabetes pathogenesis, it will be necessary to generate 3D multicellular models, which include all endocrine cell types, vasculature endothelial cells, and immune cells. When fully autologous, meaning that all cells are derived from iPSCs of a single individual with T1D and immune cells are isolated from peripheral blood of the same individual, such a 3D multicellular iPSC-islet organoid could provide a platform for studying the mechanisms of immune-mediated beta cell destruction in T1D. Similarly, the inclusion of vasculature and innate immune cells into a cell model could help define how a pro-inflammatory islet microenvironment, as observed in T2D, contributes to beta cell dysfunction and destruction (Eguchi and Nagai 2017). When introduced into a microfluidic “organ-on-a-chip” platform (Lenguito et al. 2017), 3D hPSC-islet organoids could serve as cost-efficient models for screening pharmaceutical agents, mimicking normal physiology, and studying disease.
Figure 4.
Innovative platforms for studying human beta cells in health and disease. Multiple cell types present in the pancreatic islet in vivo niche are generated from a single human iPSC line and cultured as a 3D organoid in microfluidic devices to mimic in vivo conditions. These platforms can be adapted for small molecule or genetic screens and interrogated by live imaging of fluorescent reporters or with functional and molecular assays. (hPSCs) human pluripotent stem cells.
Acknowledgments
We apologize to our colleagues whose references were omitted owing to space constraints. We thank Jenna Kovsky and Ryan Geusz for critical comments on the manuscript. Work in the Sander laboratory is supported by grants from the National Institutes of Health (R01DK068471, R01DK078803, UG3DK120005, R01DK122607), the Juvenile Diabetes Research Foundation, and the California Institute of Regenerative Medicine. B.G. was supported by a postdoctoral fellowship from the Larry L. Hillblom Foundation (2015-D-021-FEL).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.331397.119.
References
- Akerman I, Tu Z, Beucher A, Rolando DMY, Sauty-Colace C, Benazra M, Nakic N, Yang J, Wang H, Pasquali L, et al. 2017. Human pancreatic β cell lncRNAs control cell-specific regulatory networks. Cell Metab 25: 400–411. 10.1016/j.cmet.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen HL, Flanagan SE, Shaw-Smith C, De Franco E, Akerman I, Caswell R, International Pancreatic Agenesis Consortium, Ferrer J, Hattersley AT, Ellard S. 2014. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet 44: 20–22. 10.1038/ng.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S, Cook B, Zhou T, Ghazizadeh Z, Lis R, Zhang T, Khalaj M, Crespo M, Perera M, Xiang JZ, et al. 2018. Discovery of a drug candidate for GLIS3-associated diabetes. Nat Commun 9: 2681 10.1038/s41467-018-04918-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L. 2016. βlinc1 encodes a long noncoding RNA that regulates islet β-cell formation and function. Genes Dev 30: 502–507. 10.1101/gad.273821.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa D, Saarimäki-Vire J, Borshagovski D, Survila M, Lindholm P, Galli E, Eurola S, Ustinov J, Grym H, Huopio H, et al. 2018. Insulin mutations impair β-cell development in a patient-derived iPSC model of neonatal diabetes. Elife 7 10.7554/eLife.38519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas-Ponce A, Tritschler S, Dony L, Scheibner K, Tarquis-Medina M, Salinno C, Schirge S, Burtscher I, Böttcher A, Theis FJ, et al. 2019. Comprehensive single cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development 146: dev173849 10.1242/dev.173849 [DOI] [PubMed] [Google Scholar]
- Bellanné-Chantelot C, Chauveau D, Gautier JF, Dubois-Laforgue D, Clauin S, Beaufils S, Wilhelm JM, Boitard C, Noël LH, Velho G, et al. 2004. Clinical spectrum associated with hepatocyte nuclear factor-1β mutations. Ann Intern Med 140: 510–517. 10.7326/0003-4819-140-7-200404060-00009 [DOI] [PubMed] [Google Scholar]
- Ben-Yosef D, Malcov M, Eiges R. 2008. PGD-derived human embryonic stem cell lines as a powerful tool for the study of human genetic disorders. Mol Cell Endocrinol 282: 153–158. 10.1016/j.mce.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Biden TJ, Boslem E, Chu KY, Sue N. 2014. Lipotoxic endoplasmic reticulum stress, β cell failure, and type 2 diabetes mellitus. Trends Endocrinol Metab 25: 389–398. 10.1016/j.tem.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Bonnefond A, Vaillant E, Philippe J, Skrobek B, Lobbens S, Yengo L, Huyvaert M, Cavé H, Busiah K, Scharfmann R, et al. 2013. Transcription factor gene MNX1 is a novel cause of permanent neonatal diabetes in a consanguineous family. Diabetes Metab 39: 276–280. 10.1016/j.diabet.2013.02.007 [DOI] [PubMed] [Google Scholar]
- Brambillasca S, Altkrueger A, Colombo SF, Friederich A, Eickelmann P, Mark M, Borgese N, Solimena M. 2012. CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1) is a tail-anchored protein in the endoplasmic reticulum (ER) of insulinoma cells. J Biol Chem 287: 41808–41819. 10.1074/jbc.M112.376558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. 2001. Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411: 965–969. 10.1038/35082103 [DOI] [PubMed] [Google Scholar]
- Brunelle M, Nordell Markovits A, Rodrigue S, Lupien M, Jacques PE, Gévry N. 2015. The histone variant H2A.Z is an important regulator of enhancer activity. Nucleic Acids Res 43: 9742–9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwick N, Coats SA, Nakamura T, Shimamura A. 2012. Impaired ribosomal subunit association in Shwachman-Diamond syndrome. Blood 120: 5143–5152. 10.1182/blood-2012-04-420166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes LE, Wong DM, Subramaniam M, Meyer NP, Gilchrist CL, Knox SM, Tward AD, Ye CJ, Sneddon JB. 2018. Lineage dynamics of murine pancreatic development at single-cell resolution. Nat Commun 9: 3922 10.1038/s41467-018-06176-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas-Diaz FL, Osorio-Quintero C, Diaz-Miranda MA, Kishore S, Leavens K, Jobaliya C, Stanescu D, Ortiz-Gonzalez X, Yoon C, Chen CS, et al. 2019. Modeling monogenic diabetes using human ESCs reveals developmental and metabolic deficiencies caused by mutations in HNF1A. Cell Stem Cell 25: 273–289.e5. 10.1016/j.stem.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Carrasco M, Delgado I, Soria B, Martín F, Rojas A. 2012. GATA4 and GATA6 control mouse pancreas organogenesis. J Clin Invest 122: 3504–3515. 10.1172/JCI63240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarino RR, Stark A. 2018. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev 32: 202–223. 10.1101/gad.310367.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebola I, Rodríguez-Seguí SA, Cho CH, Bessa J, Rovira M, Luengo M, Chhatriwala M, Berry A, Ponsa-Cobas J, Maestro MA, et al. 2015. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat Cell Biol 17: 615–626. 10.1038/ncb3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BZ, Yu SL, Singh S, Kao LP, Tsai ZY, Yang PC, Chen BH, Shoei-Lung Li S. 2011. Identification of microRNAs expressed highly in pancreatic islet-like cell clusters differentiated from human embryonic stem cells. Cell Biol Int 35: 29–37. 10.1042/CBI20090081 [DOI] [PubMed] [Google Scholar]
- Chia CY, Madrigal P, Denil S, Martinez I, Garcia-Bernardo J, El-Khairi R, Chhatriwala M, Shepherd MH, Hattersley AT, Dunn NR, et al. 2019. GATA6 cooperates with EOMES/SMAD2/3 to deploy the gene regulatory network governing human definitive endoderm and pancreas formation. Stem Cell Reports 12: 57–70. 10.1016/j.stemcr.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion JP, Reh CS, Daniels M, Liu X, Paz VP, Ye H, Highland HM, Hanis CL, Greeley SA. 2014. Neonatal diabetes, gallbladder agenesis, duodenal atresia, and intestinal malrotation caused by a novel homozygous mutation in RFX6. Pediatr Diabetes 15: 67–72. 10.1111/pedi.12063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. 2006. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol 24: 1392–1401. 10.1038/nbt1259 [DOI] [PubMed] [Google Scholar]
- Daneshvar K, Pondick JV, Kim BM, Zhou C, York SR, Macklin JA, Abualteen A, Tan B, Sigova AA, Marcho C, et al. 2016. DIGIT is a conserved long noncoding RNA that regulates GSC expression to control definitive endoderm differentiation of embryonic stem cells. Cell Rep 17: 353–365. 10.1016/j.celrep.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vas MG, Kopp JL, Heliot C, Sander M, Cereghini S, Haumaitre C. 2015. Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Development 142: 871–882. 10.1242/dev.110759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri P, Warner JT, Minton JA, Patch AM, Ellard S, Hattersley AT, Barr S, Hawkes D, Wales JK, Gregory JW. 2011. Novel GLIS3 mutations demonstrate an extended multisystem phenotype. Eur J Endocrinol 164: 437–443. 10.1530/EJE-10-0893 [DOI] [PubMed] [Google Scholar]
- Dooley J, Tian L, Schonefeldt S, Delghingaro-Augusto V, Garcia-Perez JE, Pasciuto E, Di Marino D, Carr EJ, Oskolkov N, Lyssenko V, et al. 2016. Genetic predisposition for β cell fragility underlies type 1 and type 2 diabetes. Nat Genet 48: 519–527. 10.1038/ng.3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edghill EL, Bingham C, Ellard S, Hattersley AT. 2006. Mutations in hepatocyte nuclear factor-1β and their related phenotypes. J Med Genet 43: 84–90. 10.1136/jmg.2005.032854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi K, Nagai R. 2017. Islet inflammation in type 2 diabetes and physiology. J Clin Invest 127: 14–23. 10.1172/JCI88877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esguerra JL, Eliasson L. 2014. Functional implications of long non-coding RNAs in the pancreatic islets of Langerhans. Front Genet 5: 209 10.3389/fgene.2014.00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firdous P, Nissar K, Ali S, Ganai BA, Shabir U, Hassan T, Masoodi SR. 2018. Genetic testing of maturity-onset diabetes of the young current status and future perspectives. Front Endocrinol 9: 253 10.3389/fendo.2018.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SE, De Franco E, Lango Allen H, Zerah M, Abdul-Rasoul MM, Edge JA, Stewart H, Alamiri E, Hussain K, Wallis S, et al. 2014a. Analysis of transcription factors key for mouse pancreatic development establishes NKX2-2 and MNX1 mutations as causes of neonatal diabetes in man. Cell Metab 19: 146–154. 10.1016/j.cmet.2013.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Allen HL, De Franco E, McDonald TJ, Rajala H, Ramelius A, Barton J, et al. 2014b. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet 46: 812–814. 10.1038/ng.3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Jin L, Wang X, Luo A, Hu J, Zheng X, Tsark WM, Riggs AD, Ku HT, Huang W. 2013. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci 110: 17892–17897. 10.1073/pnas.1317397110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, Kawaguchi Y, Furuyama K, Kodama S, Horiguchi M, Kuhara T, Kawaguchi M, Terao M, Doi R, Wright CV, et al. 2008. Reduction of Ptf1a gene dosage causes pancreatic hypoplasia and diabetes in mice. Diabetes 57: 2421–2431. 10.2337/db07-1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalez MA, Carette C, Bagattin A, Chiral M, Makinistoglu MP, Garbay S, Prévost G, Madaras C, Hérault Y, Leibovici M, et al. 2016. A suppressor locus for MODY3-diabetes. Sci Rep 6: 33087 10.1038/srep33087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijsbers A, Montagut DC, Méndez-Godoy A, Altamura D, Saviano M, Siliqi D, Sánchez-Puig N. 2018. Interaction of the GTPase elongation factor like-1 with the Shwachman-Diamond syndrome protein and its missense mutations. Int J Mol Sci 19: 4012 10.3390/ijms19124012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromada J, Chabosseau P, Rutter GA. 2018. The α-cell in diabetes mellitus. Nat Rev Endocrinol 14: 694–704. 10.1038/s41574-018-0097-y [DOI] [PubMed] [Google Scholar]
- Guo M, Zhang T, Dong X, Xiang JZ, Lei M, Evans T, Graumann J, Chen S. 2017. Using hESCs to probe the interaction of the diabetes-associated genes CDKAL1 and MT1E. Cell Rep 19: 1512–1521. 10.1016/j.celrep.2017.04.070 [DOI] [PubMed] [Google Scholar]
- Haldorsen IS, Vesterhus M, Ræder H, Jensen DK, Søvik O, Molven A, Njølstad PR. 2008. Lack of pancreatic body and tail in HNF1B mutation carriers. Diabet Med 25: 782–787. 10.1111/j.1464-5491.2008.02460.x [DOI] [PubMed] [Google Scholar]
- Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. 1999. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet 23: 71–75. 10.1038/12674 [DOI] [PubMed] [Google Scholar]
- Haumaitre C, Barbacci E, Jenny M, Ott MO, Gradwohl G, Cereghini S. 2005. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc Natl Acad Sci 102: 1490–1495. 10.1073/pnas.0405776102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haumaitre C, Fabre M, Cormier S, Baumann C, Delezoide AL, Cereghini S. 2006. Severe pancreas hypoplasia and multicystic renal dysplasia in two human fetuses carrying novel HNF1β/MODY5 mutations. Hum Mol Genet 15: 2363–2375. 10.1093/hmg/ddl161 [DOI] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, St Jacques B, McMahon AP, Melton DA. 2000. Regulation of pancreas development by hedgehog signaling. Development 127: 4905–4913. [DOI] [PubMed] [Google Scholar]
- Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, Morrisey EE. 2014. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev 28: 1363–1379. 10.1101/gad.238782.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohwieler M, Illing A, Hermann PC, Mayer T, Stockmann M, Perkhofer L, Eiseler T, Antony JS, Müller M, Renz S, et al. 2017. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 66: 473–486. 10.1136/gutjnl-2016-312423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa Y, Toyoda T, Fukui K, Baden MY, Funato M, Kondo Y, Sudo T, Iwahashi H, Kishida M, Okada C, et al. 2017. Insulin-producing cells derived from ‘induced pluripotent stem cells’ of patients with fulminant type 1 diabetes: vulnerability to cytokine insults and increased expression of apoptosis-related genes. J Diabetes Investig 9: 481–493. 10.1111/jdi.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Holtzinger A, Jagan I, BeGora M, Lohse I, Ngai N, Nostro C, Wang R, Muthuswamy LB, Crawford HC, et al. 2015. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nature Med 21: 1364–1371. 10.1038/nm.3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa A, Hanafusa T. 2006. Pathogenesis of fulminant type 1 diabetes. Rev Diabet Stud 3: 169–177. 10.1900/RDS.2006.3.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings RE, Berry AA, Kirkwood-Wilson R, Roberts NA, Hearn T, Salisbury RJ, Blaylock J, Piper Hanley K, Hanley NA. 2013. Development of the human pancreas from foregut to endocrine commitment. Diabetes 62: 3514–3522. 10.2337/db12-1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings RE, Berry AA, Strutt JP, Gerrard DT, Hanley NA. 2015. Human pancreas development. Development 142: 3126–3137. 10.1242/dev.120063 [DOI] [PubMed] [Google Scholar]
- Jennings RE, Berry AA, Gerrard DT, Wearne SJ, Strutt J, Withey S, Chhatriwala M, Piper Hanley K, Vallier L, Bobola N, et al. 2017. Laser capture and deep sequencing reveals the transcriptomic programmes regulating the onset of pancreas and liver differentiation in human embryos. Stem Cell Reports 9: 1387–1394. 10.1016/j.stemcr.2017.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Liu Y, Liu R, Zhang K, Zhang Y. 2015. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep 11: 137–148. 10.1016/j.celrep.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. 2007. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21: 1519–1529. 10.1101/gad.1547707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Mulas F, Gaertner B, Sui Y, Wang J, Zeng C, Vinckier N, Wang A, Nguyen-Ngoc K-V, Chiou J, et al. 2019. A network of microRNAs acts to promote cell cycle exit and differentiation of human pancreatic endocrine cells. bioRxiv doi:10.1101/618330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, Grapin-Botton A. 2007. Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev Cell 12: 457–465. 10.1016/j.devcel.2007.02.010 [DOI] [PubMed] [Google Scholar]
- Kang HS, Kim YS, ZeRuth G, Beak JY, Gerrish K, Kilic G, Sosa-Pineda B, Jensen J, Foley J, Jetten AM. 2009. Transcription factor Glis3, a novel critical player in the regulation of pancreatic β-cell development and insulin gene expression. Mol Cell Biol 29: 6366–6379. 10.1128/MCB.01259-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. 2007. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol 5: e203 10.1371/journal.pbio.0050203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krentz NAJ, Lee MYY, Xu EE, Sproul SLJ, Maslova A, Sasaki S, Lynn FC. 2018. Single-cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors. Stem Cell Reports 11: 1551–1564. 10.1016/j.stemcr.2018.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, et al. 2008. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol 26: 443–452. 10.1038/nbt1393 [DOI] [PubMed] [Google Scholar]
- Lee K, Cho H, Rickert RW, Li QV, Pulecio J, Leslie CS, Huangfu D. 2019. FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Rep 28: 382–393.e7. 10.1016/j.celrep.2019.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenguito G, Chaimov D, Weitz JR, Rodriguez-Diaz R, Rawal SA, Tamayo-Garcia A, Caicedo A, Stabler CL, Buchwald P, Agarwal A. 2017. Resealable, optically accessible, PDMS-free fluidic platform for ex vivo interrogation of pancreatic islets. Lab Chip 17: 772–781. 10.1039/C6LC01504B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Arber S, Jessell TM, Edlund H. 1999. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat Genet 23: 67–70. 10.1038/12669 [DOI] [PubMed] [Google Scholar]
- Li J, Wu X, Zhou Y, Lee M, Guo L, Han W, Mo W, Cao WM, Sun D, Xie R, et al. 2018. Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Res 46: 2883–2900. 10.1093/nar/gky063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QV, Dixon G, Verma N, Rosen BP, Gordillo M, Luo R, Xu C, Wang Q, Soh CL, Yang D, et al. 2019. Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat Genet 51: 999–1010. 10.1038/s41588-019-0408-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Zhang Y, Han J, Wang W, Liu Y, Li J, Jiang X. 2015. Embryonic stem cell-derived pancreatic endoderm transplant with MCT1-suppressing miR-495 attenuates type II diabetes in mice. Endocr J 62: 907–920. 10.1507/endocrj.EJ15-0186 [DOI] [PubMed] [Google Scholar]
- Liao X, Xue H, Wang YC, Nazor KL, Guo S, Trivedi N, Peterson SE, Liu Y, Loring JF, Laurent LC. 2013. Matched miRNA and mRNA signatures from an hESC-based in vitro model of pancreatic differentiation reveal novel regulatory interactions. J Cell Sci 126: 3848–3861. 10.1242/jcs.123570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. 1999. Requirement for Wnt3 in vertebrate axis formation. Nat Genet 22: 361–365. 10.1038/11932 [DOI] [PubMed] [Google Scholar]
- Liu M, Hodish I, Haataja L, Lara-Lemus R, Rajpal G, Wright J, Arvan P. 2010. Proinsulin misfolding and diabetes: mutant INS gene-induced diabetes of youth. Trends Endocrinol Metab 21: 652–659. 10.1016/j.tem.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, Lee KL, Choo SH, Lim CY, Nichane M, et al. 2014. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell 14: 237–252. 10.1016/j.stem.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, et al. 2016. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18: 637–652. 10.1016/j.stem.2016.01.024 [DOI] [PubMed] [Google Scholar]
- Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. 2007. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes 56: 2938–2945. 10.2337/db07-0175 [DOI] [PubMed] [Google Scholar]
- Maehr R, Chen S, Snitow M, Ludwig T, Yagasaki L, Goland R, Leibel RL, Melton DA. 2009. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci 106: 15768–15773. 10.1073/pnas.0906894106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N, et al. 2018. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 50: 1505–1513. 10.1038/s41588-018-0241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi A, Prawiro C, Seymour PA, de Lichtenberg KH, Jackson A, Serup P, Semb H. 2018. Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature 564: 114–118. 10.1038/s41586-018-0762-2 [DOI] [PubMed] [Google Scholar]
- Manzar GS, Kim EM, Zavazava N. 2017. Demethylation of induced pluripotent stem cells from type 1 diabetic patients enhances differentiation into functional pancreatic β cells. J Biol Chem 292: 14066–14079. 10.1074/jbc.M117.784280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Rutter GA, Latreille M. 2016. MiRNAs in β-cell development, identity, and disease. Front Genet 7: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury Y, Gauthier M, Peschanski M, Martinat C. 2012. Human pluripotent stem cells for disease modelling and drug screening. Bioessays 34: 61–71. 10.1002/bies.201100071 [DOI] [PubMed] [Google Scholar]
- McGrath PS, Watson CL, Ingram C, Helmrath MA, Wells JM. 2015. The basic helix-loop-helix transcription factor NEUROG3 is required for development of the human endocrine pancreas. Diabetes 64: 2497–2505. 10.2337/db14-1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi Y, Guo N, He T, Ji J, Li Z, Huang P. 2015. miR-410 enhanced hESC-derived pancreatic endoderm transplant to alleviate gestational diabetes mellitus. J Mol Endocrinol 55: 219–229. 10.1530/JME-15-0100 [DOI] [PubMed] [Google Scholar]
- Millman JR, Xie C, Van Dervort A, Gürtler M, Pagliuca FW, Melton DA. 2016. Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat Commun 7: 11463 10.1038/ncomms11463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakić N, García-Hurtado J, Rodríguez-Seguí S, et al. 2012. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab 16: 435–448. 10.1016/j.cmet.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motterle A, Gattesco S, Peyot ML, Esguerra JLS, Gomez-Ruiz A, Laybutt DR, Gilon P, Burdet F, Ibberson M, Eliasson L, et al. 2017. Identification of islet-enriched long non-coding RNAs contributing to β-cell failure in type 2 diabetes. Mol Metab 6: 1407–1418. 10.1016/j.molmet.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair G, Hebrok M. 2015. Islet formation in mice and men: lessons for the generation of functional insulin-producing β-cells from human pluripotent stem cells. Curr Opin Genet Dev 32: 171–180. 10.1016/j.gde.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair GG, Liu JS, Russ HA, Tran S, Saxton MS, Chen R, Juang C, Li ML, Nguyen VQ, Giacometti S, et al. 2019. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nat Cell Biol 21: 263–274. 10.1038/s41556-018-0271-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng NHJ, Jasmen JB, Lim CS, Lau HH, Krishnan VG, Kadiwala J, Kulkarni RN, Ræder H, Vallier L, Hoon S, et al. 2019. HNF4A haploinsufficiency in MODY1 abrogates liver and pancreas differentiation from patient-derived induced pluripotent stem cells. iScience 16: 192–205. 10.1016/j.isci.2019.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Q, Gu Y, Xie Y, Yin Q, Zhang H, Nie A, Li W, Wang Y, Ning G, Wang W, et al. 2017. Raptor regulates functional maturation of murine β cells. Nat Commun 8: 15755 10.1038/ncomms15755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Hevia P, Garcia E, Klein D, Alvarez-Cubela S, Bravo-Egana V, Rosero S, Damaris Molano R, Vargas N, Ricordi C, et al. 2012. Antisense miR-7 impairs insulin expression in developing pancreas and in cultured pancreatic buds. Cell Transplant 21: 1761–1774. 10.3727/096368911X612521 [DOI] [PubMed] [Google Scholar]
- O'Brien J, Hayder H, Zayed Y, Peng C. 2018. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol 9: 402 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. 1996. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122: 983–995. [DOI] [PubMed] [Google Scholar]
- Otonkoski T, Roivainen M, Vaarala O, Dinesen B, Leipälä JA, Hovi T, Knip M. 2000. Neonatal type I diabetes associated with maternal echovirus 6 infection: a case report. Diabetologia 43: 1235–1238. 10.1007/s001250051518 [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. 2014. Generation of functional human pancreatic β cells in vitro. Cell 159: 428–439. 10.1016/j.cell.2014.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES. 2018. Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front Genet 9: 440 10.3389/fgene.2018.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, Mularoni L, Miguel-Escalada I, Akerman I, Tena JJ, Morán I, Gómez-Marin C, van de Bunt M, et al. 2014. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 46: 136–143. 10.1038/ng.2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen MBK, Azad A, Ingvorsen C, Hess K, Hansson M, Grapin-Botton A, Honoré C. 2017. Single-cell gene expression analysis of a human ESC model of pancreatic endocrine development reveals different paths to β-cell differentiation. Stem Cell Reports 9: 1246–1261. 10.1016/j.stemcr.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoglio M, Sreenan S, Roe M, Pugh W, Ostrega D, Doyen A, Pick AJ, Baldwin A, Velho G, Froguel P, et al. 1998. Defective insulin secretion in hepatocyte nuclear factor 1α-deficient mice. J Clin Invest 101: 2215–2222. 10.1172/JCI2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramond C, Glaser N, Berthault C, Ameri J, Kirkegaard JS, Hansson M, Honoré C, Semb H, Scharfmann R. 2017. Reconstructing human pancreatic differentiation by mapping specific cell populations during development. Elife 6 10.7554/eLife.27564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramond C, Beydag-Tasöz BS, Azad A, van de Bunt M, Petersen MBK, Beer NL, Glaser N, Berthault C, Gloyn AL, Hansson M, et al. 2018. Understanding human fetal pancreas development using subpopulation sorting, RNA sequencing and single-cell profiling. Development 145: dev165480 10.1242/dev.165480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, Riedel MJ, Wideman RD, Karanu F, Ao Z, Warnock GL, Kieffer TJ. 2011. Production of functional glucagon-secreting α-cells from human embryonic stem cells. Diabetes 60: 239–247. 10.2337/db10-0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O'Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. 2014. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol 32: 1121–1133. 10.1038/nbt.3033 [DOI] [PubMed] [Google Scholar]
- Romer AI, Sussel L. 2015. Pancreatic islet cell development and regeneration. Curr Opin Endocrinol Diabetes Obes 22: 255–264. 10.1097/MED.0000000000000174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado-Olivieri EA, Anderson K, Kenty JH, Melton DA. 2019. YAP inhibition enhances the differentiation of functional stem cell-derived insulin-producing β cells. Nat Commun 10: 1464 10.1038/s41467-019-09404-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BL, Zaret KS. 2001. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev 15: 1998–2009. 10.1101/gad.904601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Cabezas O, Minton JA, Kantor I, Williams D, Ellard S, Hattersley AT. 2010. Homozygous mutations in NEUROD1 are responsible for a novel syndrome of permanent neonatal diabetes and neurological abnormalities. Diabetes 59: 2326–2331. 10.2337/db10-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Cabezas O, Jensen JN, Hodgson MI, Codner E, Ellard S, Serup P, Hattersley AT. 2011. Permanent neonatal diabetes and enteric anendocrinosis associated with biallelic mutations in NEUROG3. Diabetes 60: 1349–1353. 10.2337/db10-1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ HA, Parent AV, Ringler JJ, Hennings TG, Nair GG, Shveygert M, Guo T, Puri S, Haataja L, Cirulli V, et al. 2015. Controlled induction of human pancreatic progenitors produces functional β-like cells in vitro. EMBO J 34: 1759–1772. 10.15252/embj.201591058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarimäki-Vire J, Balboa D, Russell MA, Saarikettu J, Kinnunen M, Keskitalo S, Malhi A, Valensisi C, Andrus C, Eurola S, et al. 2017. An activating STAT3 mutation causes neonatal diabetes through premature induction of pancreatic differentiation. Cell Rep 19: 281–294. 10.1016/j.celrep.2017.03.055 [DOI] [PubMed] [Google Scholar]
- Saunders DC, Brissova M, Phillips N, Shrestha S, Walker JT, Aramandla R, Poffenberger G, Flaherty DK, Weller KP, Pelletier J et al. 2018. Ectonucleoside triphosphate diphosphohydrolase-3 antibody targets adult human pancreatic β cells for in vitro and in vivo analysis. Cell Metab 29: 745–754.E4. 10.1016/j.cmet.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavuzzo MA, Hill MC, Chmielowiec J, Yang D, Teaw J, Sheng K, Kong Y, Bettini M, Zong C, Martin JF, et al. 2018. Endocrine lineage biases arise in temporally distinct endocrine progenitors during pancreatic morphogenesis. Nat Commun 9: 3356 10.1038/s41467-018-05740-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick GS, Barker KT, Stolte-Dijkstra I, Fleischmann C, Coleman RJ, Garrett C, Gloyn AL, Edghill EL, Hattersley AT, Wellauer PK, et al. 2004. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet 36: 1301–1305. 10.1038/ng1475 [DOI] [PubMed] [Google Scholar]
- Senée V, Chelala C, Duchatelet S, Feng D, Blanc H, Cossec JC, Charon C, Nicolino M, Boileau P, Cavener DR, et al. 2006. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet 38: 682–687. 10.1038/ng1802 [DOI] [PubMed] [Google Scholar]
- Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. 2007. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci 104: 1865–1870. 10.1073/pnas.0609217104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour PA, Shih HP, Patel NA, Freude KK, Xie R, Lim CJ, Sander M. 2012. A Sox9/Fgf feed-forward loop maintains pancreatic organ identity. Development 139: 3363–3372. 10.1242/dev.078733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L, Hua H, Foo K, Martinez H, Watanabe K, Zimmer M, Kahler DJ, Freeby M, Chung W, LeDuc C, et al. 2014. β-cell dysfunction due to increased ER stress in a stem cell model of Wolfram syndrome. Diabetes 63: 923–933. 10.2337/db13-0717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N, Chawla R, Mueller J, Vanderhooft J, Whitehorn LJ, Rosenthal B, Gürtler M, Estanboulieh RR, Shvartsman D, Gifford DK, et al. 2019a. A peninsular structure coordinates asynchronous differentiation with morphogenesis to generate pancreatic islets. Cell 176: 790–804.e13. 10.1016/j.cell.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N, Vanderhooft J, Straubhaar J, Mueller J, Chawla R, Zhou Q, Engquist EN, Trapnell C, Gifford DK, Melton DA. 2019b. Wnt signaling separates the progenitor and endocrine compartments during pancreas development. Cell Rep 27: 2281–2291.e5. 10.1016/j.celrep.2019.04.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi ZD, Lee K, Yang D, Amin S, Verma N, Li QV, Zhu Z, Soh CL, Kumar R, Evans T, et al. 2017. Genome editing in hPSCs reveals GATA6 haploinsufficiency and a genetic interaction with GATA4 in human pancreatic development. Cell Stem Cell 20: 675–688.e6. 10.1016/j.stem.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Wang A, Sander M. 2013. Pancreas organogenesis: from lineage determination to morphogenesis. Annu Rev Cell Dev Biol 29: 81–105. 10.1146/annurev-cellbio-101512-122405 [DOI] [PubMed] [Google Scholar]
- Shih HP, Seymour PA, Patel NA, Xie R, Wang A, Liu PP, Yeo GW, Magnuson MA, Sander M. 2015. A gene regulatory network cooperatively controlled by Pdx1 and Sox9 governs lineage allocation of foregut progenitor cells. Cell Rep 13: 326–336. 10.1016/j.celrep.2015.08.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, et al. 2013. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci 110: 2876–2881. 10.1073/pnas.1221904110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek S, Zhou T, Robinson CL, Tsai SY, Crespo M, Amin S, Lin X, Hon J, Evans T, Chen S. 2016. Modeling cystic fibrosis using pluripotent stem cell-derived human pancreatic ductal epithelial cells. Stem Cells Transl Med 5: 572–579. 10.5966/sctm.2015-0276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinagoga KL, Stone WJ, Schiesser JV, Schweitzer JI, Sampson L, Zheng Y, Wells JM. 2017. Distinct roles for the mTOR pathway in postnatal morphogenesis, maturation and function of pancreatic islets. Development 144: 2402–2414. 10.1242/dev.146316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Qu HQ, Taleb N, Kishimoto NY, Scheel DW, Lu Y, Patch AM, Grabs R, Wang J, Lynn FC, et al. 2010. Rfx6 directs islet formation and insulin production in mice and humans. Nature 463: 775–780. 10.1038/nature08748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Staffers DA, Ferrer J, Clarke WL, Habener JF. 1997. Early-onset type-ll diabetes mellitus (MODY4) linked to IPF1. Nat Genet 17: 138–139. 10.1038/ng1097-138 [DOI] [PubMed] [Google Scholar]
- Stephenson EL, Mason C, Braude PR. 2009. Preimplantation genetic diagnosis as a source of human embryonic stem cells for disease research and drug discovery. BJOG 116: 158–165. 10.1111/j.1471-0528.2008.02009.x [DOI] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. 2013. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154: 888–903. 10.1016/j.cell.2013.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel M, Duncan SA. 1997. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4α regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci 94: 13209–13214. 10.1073/pnas.94.24.13209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. 1997. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet 15: 106–110. 10.1038/ng0197-106 [DOI] [PubMed] [Google Scholar]
- Sui L, Danzl N, Campbell SR, Viola R, Williams D, Xing Y, Wang Y, Phillips N, Poffenberger G, Johannesson B, et al. 2018. β-cell replacement in mice using human type 1 diabetes nuclear transfer embryonic stem cells. Diabetes 67: 26–35. 10.2337/db17-0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa-Shirasawa S, Yoshie S, Yue F, Mogi A, Yokoyama T, Tomotsune D, Sasaki K. 2013. FGF7 and cell density are required for final differentiation of pancreatic amylase-positive cells from human ES cells. Cell Tissue Res 354: 751–759. 10.1007/s00441-013-1695-6 [DOI] [PubMed] [Google Scholar]
- Teo AK, Tsuneyoshi N, Hoon S, Tan EK, Stanton LW, Wright CV, Dunn NR. 2015. PDX1 binds and represses hepatic genes to ensure robust pancreatic commitment in differentiating human embryonic stem cells. Stem Cell Reports 4: 578–590. 10.1016/j.stemcr.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo AK, Lau HH, Valdez IA, Dirice E, Tjora E, Raeder H, Kulkarni RN. 2016. Early developmental perturbations in a human stem cell model of MODY5/HNF1B pancreatic hypoplasia. Stem Cell Reports 6: 357–367. 10.1016/j.stemcr.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatava T, Kudva YC, Edukulla R, Squillace K, De Lamo JG, Khan YK, Sakuma T, Ohmine S, Terzic A, Ikeda Y. 2013. Intrapatient variations in type 1 diabetes-specific iPS cell differentiation into insulin-producing cells. Mol Ther 21: 228–239. 10.1038/mt.2012.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen SK, Gloyn AL. 2014. The pancreatic β cell: recent insights from human genetics. Trends Endocrinol Metab 25: 425–434. 10.1016/j.tem.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyaboonchai A, Cardenas-Diaz FL, Ying L, Maguire JA, Sim X, Jobaliya C, Gagne AL, Kishore S, Stanescu DE, Hughes N, et al. 2017. GATA6 plays an important role in the induction of human definitive endoderm, development of the pancreas, and functionality of pancreatic β cells. Stem Cell Reports 8: 589–604. 10.1016/j.stemcr.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropel P, Tournois J, Côme J, Varela C, Moutou C, Fragner P, Cailleret M, Laâbi Y, Peschanski M, Viville S. 2010. High-efficiency derivation of human embryonic stem cell lines following pre-implantation genetic diagnosis. In vitro Cell Dev Biol Anim 46: 376–385. 10.1007/s11626-010-9300-8 [DOI] [PubMed] [Google Scholar]
- Tulpule A, Kelley JM, Lensch MW, McPherson J, Park IH, Hartung O, Nakamura T, Schlaeger TM, Shimamura A, Daley GQ. 2013. Pluripotent stem cell models of Shwachman-Diamond syndrome reveal a common mechanism for pancreatic and hematopoietic dysfunction. Cell Stem Cell 12: 727–736. 10.1016/j.stem.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazco-Cruz L, Song J, Maxwell KG, Goedegebuure MM, Augsornworawat P, Hogrebe NJ, Millman JR. 2019. Acquisition of dynamic function in human stem cell-derived β cells. Stem Cell Reports 12: 351–365. 10.1016/j.stemcr.2018.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Workman JL. 2015. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol 16: 178–189. 10.1038/nrm3941 [DOI] [PubMed] [Google Scholar]
- Veres A, Faust AL, Bushnell HL, Engquist EN, Kenty JH, Harb G, Poh YC, Sintov E, Gürtler M, Pagliuca FW, et al. 2019. Charting cellular identity during human in vitro β-cell differentiation. Nature 569: 368–373. 10.1038/s41586-019-1168-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethe H, Bjørlykke Y, Ghila LM, Paulo JA, Scholz H, Gygi SP, Chera S, Ræder H. 2017. Probing the missing mature β-cell proteomic landscape in differentiating patient iPSC-derived cells. Sci Rep 7: 4780 10.1038/s41598-017-04979-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal JA, Ventura A. 2015. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol 25: 137–147. 10.1016/j.tcb.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, Muth K, Palmer J, Qiu Y, Wang J, et al. 2015. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 16: 386–399. 10.1016/j.stem.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sterr M, Ansarullah, Burtscher I, Böttcher A, Beckenbauer J, Siehler J, Meitinger T, Häring HU, Staiger H, et al. 2019. Point mutations in the PDX1 transactivation domain impair human β-cell development and function. Mol Metab 24: 80–97. 10.1016/j.molmet.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CR, O'Sullivan JF, Friesen M, Becker CE, Zhang X, Liu P, Wakabayashi Y, Morningstar JE, Shi X, Choi J, et al. 2017. Induced pluripotent stem cell differentiation enables functional validation of GWAS variants in metabolic disease. Cell Stem Cell 20: 547–557.e7. 10.1016/j.stem.2017.01.010 [DOI] [PubMed] [Google Scholar]
- Weedon MN, Cebola I, Patch AM, Flanagan SE, De Franco E, Caswell R, Rodríguez-Seguí SA, Shaw-Smith C, Cho CH, Allen HL, et al. 2014. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat Genet 46: 61–64. 10.1038/ng.2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Everett LJ, Lim HW, Patel NA, Schug J, Kroon E, Kelly OG, Wang A, D'Amour KA, Robins AJ, et al. 2013. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell 12: 224–237. 10.1016/j.stem.2012.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CR, Li LC, Donahue G, Ying L, Zhang YW, Gadue P, Zaret KS. 2014. Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. EMBO J 33: 2157–2170. 10.15252/embj.201488671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan S, Borok MJ, Decker KJ, Battle MA, Duncan SA, Hale MA, Macdonald RJ, Sussel L. 2012. Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. J Clin Invest 122: 3516–3528. 10.1172/JCI63352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegre Amorim M, Houghton JA, Carmo S, Salva I, Pita A, Pereira-da-Silva L. 2015. Mitchell-Riley syndrome: a novel mutation in RFX6 gene. Case Rep Genet 2015: 1–3. 10.1155/2015/937201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Guo M, Zhou T, Tan L, Chong CN, Zhang T, Dong X, Xiang JZ, Yu AS, Yue L, et al. 2016. An isogenic human ESC platform for functional evaluation of genome-wide-association-study-identified diabetes genes and drug discovery. Cell Stem Cell 19: 326–340. 10.1016/j.stem.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Li QV, Lee K, Rosen BP, González F, Soh CL, Huangfu D. 2016. Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. Cell Stem Cell 18: 755–768. 10.1016/j.stem.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]