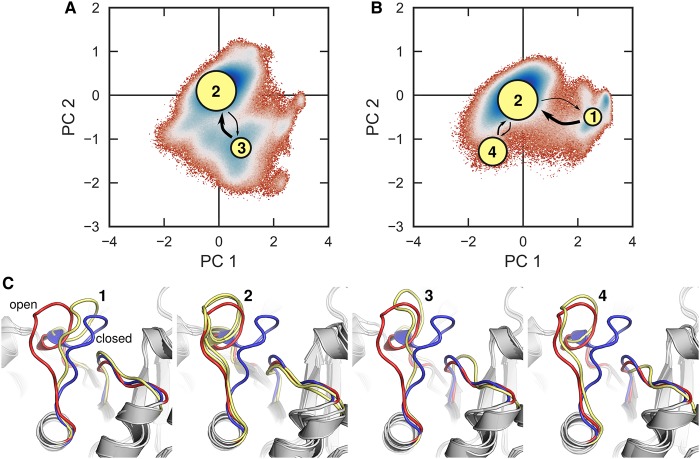
Figure 5. Modeling the open and closed conformations of TIM.
Markov state models (MSM) of (A) substrate-free TIM and (B) TIM in complex with substrate DHAP, superimposed on free energy surfaces defined in terms of the first two principal components, PC1 and PC2, obtained from conventional molecular dynamics simulations of each system (60 independent simulations, to a total of 22 μs of simulation time). The population of each node corresponding to a metastable state is represented through the area of the node, while the thickness of the arrows connecting the node corresponds to the transition probabilities, which are shown in the Supporting Information of ref. [45]. (C) A comparison of the crystal structures for TIM with open (red, PDB ID: 1YPI [71,81]) and closed (blue, PDB ID: 1NEY [80,81]) conformations of loop 6, as well as representative structures from the different centroids shown in panels (A,B). The numbering corresponds to the numbering of the different centroids. It can be seen here that the substrate-bound enzyme samples a semi-closed conformation (1), both systems sample an open conformation that is common to the two systems (2), while each samples a third open conformation that is unique to each system [(3) in the case of the substrate-free enzyme, (4) in the case of the substrate-bound enzyme]. For simulation details, see ref. [45]. This figure was originally presented in ref. [45] (https://pubs.acs.org/doi/pdf/10.1021/jacs.8b09378), and is reproduced with permission from the American Chemical Society. Please note that requests for permissions regarding further reuse of this figure should be directed to the American Chemical Society.