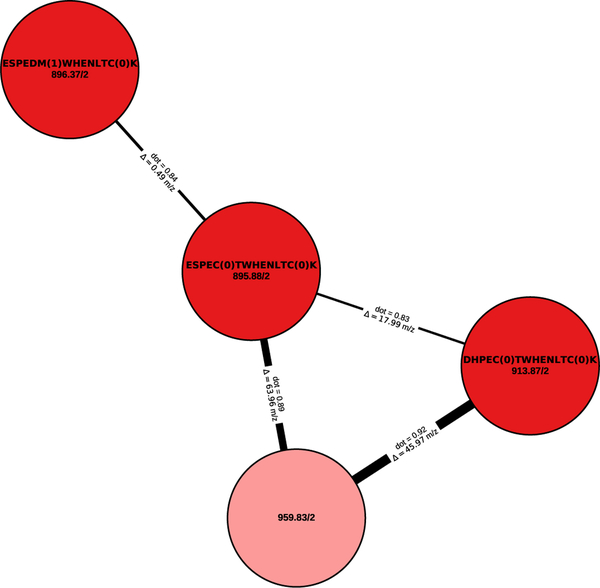
Figure 3:
A spectral network connects (un)modified spectra. The peptide sequence (if known) and the precursor mass and precursor charge are shown for each node in the spectral network. Edges between two nodes are annotated with the corresponding precursor mass difference. The spectral similarity based on the shifted dot product is indicated by the weight of the edge.
The spectral network shows a strong similarity between multiple spectra despite small differences in the identified sequences due to amino acid substitutions (CT ↔ DM, ES ↔ DH). Although the spectrum corresponding to the light red shaded node could not be fully identified through de novo searching, its high similarity to related spectra indicates that it was likely derived from the same peptide. Indeed, a full identification was precluded by the absence of any successfully matched b-ions, while the C-terminal tag “CTWHENLTCK” could still be annotated based on the y-ions.