Abstract
As computational capabilities have advanced, radiologists and their collaborators have looked for novel ways to analyze diagnostic images. This has resulted in the development of radiomics and radiogenomics as new fields in medical imaging. Radiomics and radiogenomics may change the practice of medicine, particularly for patients with colorectal cancer. Radiomics corresponds to the extraction and analysis of numerous quantitative imaging features from conventional imaging modalities in correlation with several endpoints, including the prediction of pathology, genomics, therapeutic response, and clinical outcome. In radiogenomics, qualitative and/or quantitative imaging features are extracted and correlated with genetic profiles of the imaged tissue. Thus far, several studies have evaluated the use of radiomics and radiogenomics in patients with colorectal cancer; however, there are challenges to be overcome before its routine implementation including challenges related to sample size, model design and interpretability, and the lack of robust multicenter validation set. In this article, we will review the concepts of radiomics and radiogenomics and their potential applications in rectal cancer. Radiologists should be aware of the basic concepts, benefits, pitfalls and limitations of new radiomic and radiogenomics techniques to achieve a balanced interpretation of the results.
Keywords: Radiomics, Radiogenomics, Rectal Neoplasms, Magnetic Resonance Imaging, Computed Tomography, Positron Emission Tomography
1. Introduction
As computational capabilities have advanced, radiologists and their collaborators have looked for novel ways to analyze diagnostic images. This has resulted in the development of radiomics and radiogenomics as new fields in medical imaging.
Radiomics involves the selection and segmentation of an area of interest on imaging, and subsequently the high-throughput extraction of quantitative data from the area of interest using a computer program (1). The quantitative data is analyzed with a specific clinical question in mind, such as correlation with tumor histology or genetic variants (2), classification of benign and malignant tissue (3), prediction of response to therapy prior to initiation (4, 5), and assessment of response to therapy following its completion (6).
In the current era of personalized medicine in oncology, while genetic and molecular analysis improves our ability to predict which treatment will work best for a specific patient, it comes at a high cost and has limited availability. In this context, the concept of radiogenomics which involves the identification of qualitative and/or quantitative imaging phenotypes that are characteristic of a specific genetic profile has the potential to significantly improve decision making in treatment selection and thereby improve patient outcomes (8, 9).
Radiomics and radiogenomics can dramatically change the practice of medicine, particularly in oncology. In this article, we will review the concepts of radiomics and radiogenomics and their potential applications in rectal cancer.
2. Radiomics
a. General concepts
i. Definition
Radiomics corresponds to the extraction and analysis of numerous quantitative imaging features from conventional imaging modalities in correlation with several endpoints, including the prediction of pathology, genomics, therapeutic response, and clinical outcome (1).
ii. Workflow process
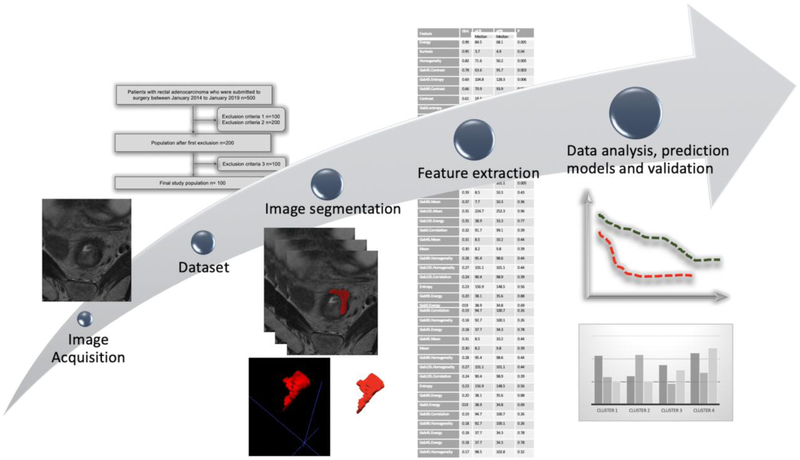
The radiomics workflow encompasses the following steps: (a) image acquisition; (b) creation of a dataset with appropriate clinical and radiological data; (c) export of DICOM images from PACS (Picture Archiving and Communication System) to the computer software that will be used to perform the radiomics analysis; (d) image segmentation of the region of interest (ROI), which corresponds to one slide of the image or volume of interest (VOI), the volume of a specific area; (e) feature extraction; (f) selection of the most relevant features to generate the appropriate model to predict the chosen endpoint; and (g) validation of the model with an internal and/or external dataset (1). Figure 1 summarizes the main steps in the radiomics workflow.
Fig. 1.
Radiomics workflow.
During imaging acquisition and dataset creation, it is important to identify the appropriate clinical variables and include high-quality imaging exams. To develop and validate radiomic models, large datasets are needed. As variations in the imaging protocol may cause differences in the textural features unrelated to biological changes and consequently impair the accuracy of the algorithm (11), it is important to include exams with similar reconstruction algorithms. The use of correction and calibration algorithms may be helpful to enhance retrospective datasets (12).
Frequently, the selected imaging exams are exported from the PACS to a radiomics software program for analysis. Several software packages are available for radiomics analysis, some as a free open source tool and others that are sold commercially (12).
Image segmentation of the target ROI or VOI (the entire volume of the area is preferable) can be done manually, automatically or semi-automatically. There are advantages and disadvantages to each approach. Automatic and semi-automatic segmentation methods have higher repeatability but are not as precise as manual segmentation in some situations, e.g., delineating the rectal tumor bed after neoadjuvant therapy. On the other hand, manual segmentation is prone to inter-reader variability and is much more time consuming (13). There is ongoing research to develop improved segmentation tools that can provide fast, accurate, and less time-consuming workflows, which will significantly improve radiomics research.
iii. Feature extraction and categories
Quantitative information derived from the voxels of segmented images are called “features.” A high number of features can be extracted from segmented images. Following feature extraction, the features can be evaluated in different ways and grouped into different categories.
Morphological features depict the size, shape and location of the segmented area (12). Textural features evaluate the distribution and relationships of the pixels or voxels. Several methods can be used to evaluate textural features: (a) statistical, which evaluates the distribution of the grayscale values; (b) model-based, which evaluates the irregularity of the area; and (c) transform-based, which transforms a spatial information into frequency (14).
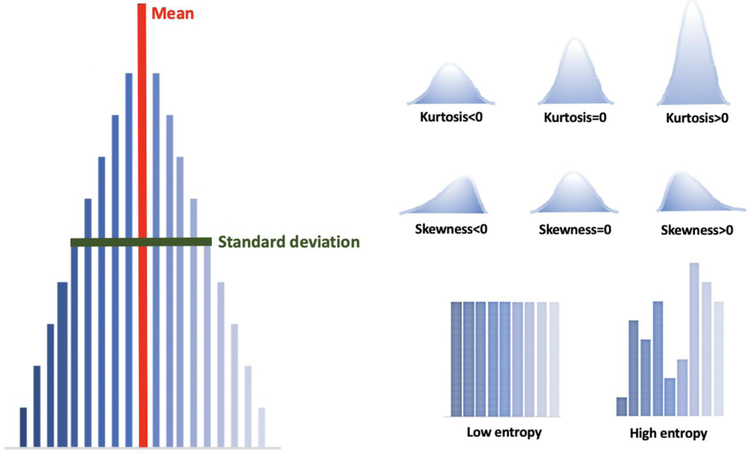
Of these, the statistical method is most commonly used, and the number of stages required to obtain the quantitative information in that model determines the “order” (14). First-order statistical textural analysis is based on an intensity histogram that takes into account the pixel values, regardless of their interactions with the neighboring pixel values, and provides no information on the spatial distribution of pixels (12, 14, 15). The most common first order parameters (Figure 2) are:
Fig. 2.
First-order statistical textural features.
Mean: average intensity of the pixels in the ROI/VOI. The higher the mean, the whiter the image.
Standard deviation: dispersion from the mean. High values indicate a wide range in pixel intensity across the ROI/VOI.
Skewness: asymmetry of histogram It measures the asymmetry of the distribution of the values in the image comparing to the mean of the values. Positive skewness indicates that the right side of the histogram is longer than the left, while negative skewness indicates that the left side is longer than the right.
Kurtosis: magnitude of pixel distribution. Positive kurtosis indicates that the histogram is taller than the normal distribution, and negative kurtosis indicates that the histogram is flatter than the normal distribution.
Entropy: irregularity of the structure. High values indicate a more heterogeneous area, while low values indicate a more homogeneous area (16).
Second-order statistical texture analysis considers the spatial relationship between 2 pixels. The second-order statistical features are classified into 3 classes: (a) grey level co-occurrence matrix (GLCM), (b) run length matrix (RLM) and (c) grey level size zone matrix (GLSZM). GLCM takes into account the frequency of specific gray values along a distance or direction, RLM takes into account the length of consecutive pixels or voxels with the same grey values in a specific direction, and GLSZM takes into account the length of consecutive pixels or voxels with the same grey values in all directions.
Superior order statistical texture analysis takes into account the neighborhood gray difference matrices and the relationship between 3 or more pixels (12, 14, 15).
Artificial intelligence and machine learning can be helpful in the evaluation of radiomic features. Deep learning convolutional neural networks can perform massive texture analysis and training of the data to create prediction algorithms. In addition, clinical and pathological variables can also be included in radiomic models to enhance their accuracy (12). After the construction of a model, validation is a final and extremely relevant step to better assess the performance of the model. Validation can be performed with internal or external datasets.
iv. Limitations and challenges to be overcome
There are several limitations and challenges to overcome before radiomics can be implemented into clinical routine. Protocol and scanner variations between different institutions and even within the same institution can reduce the robustness of the models. To date, radiomic studies that have been published have featured variations in the methodology used for texture extraction and model interpretation. Furthermore, numerous features can be extracted, and frequently the number of features is larger than the study population, which can lead to overfitting of a radiomic model. Lastly, unbalanced data is frequent in clinical practice and may necessitate the use of oversampling techniques which may possibly artificially improve the performance of the radiomic model (17, 18).
Internal and especially external validation is of key importance to assess the accuracy of radiomic models. However, data sharing among institutions is met with patient privacy challenges and can be a limiting factor. Further studies are necessary to overcome these limitations. The use of a standard imaging protocol, reproducible and consistent segmentation processes, and collaboration among various institutions to create a large annotated dataset will facilitate the establishment of reliable radiomic models.
b. Rectal applications
Several studies are already exploring the potential applications of radiomics in colorectal cancer (CRC). These applications can be categorized into the following groups: (a) evaluation of the repeatability of textural features; (b) prediction of pathological complete response after neoadjuvant chemoradiotherapy (CRT) in locally advanced rectal cancer (LARC); (d) prediction of other pathological features; (c) prediction of survival; (e) prediction of genetic profile (radiogenomics).
i. Repeatability of textural features
Several studies have evaluated the inter- and/or intra-reader agreement among radiomic features in their population and have demonstrated high values of concordance (Tables 1-3) (28, 36-38, 43-47). Few studies have been performed to specifically assess the repeatability of texture features in CRC. Badic et al. showed that textural features obtained from contrast and non-contrast enhanced CT were not equivalent, and they concluded that both images should be included when available (48). Gourtsoyianni et al. conducted a study with a small sample size on the repeatability of MRI texture features in rectal cancer on MRI and showed that first order textural features and fractal parameters had higher repeatability than high order parameters (47). Horvat et al. demonstrated that inter-reader agreement was not significantly different between readers when comparing different magnetic field strengths (1.5T vs 3T) (25).
Table 1.
Characteristics of studies that have evaluated radiomics in predicting pathological complete response after chemoradiotherapy in patients with locally advanced rectal cancer.
| Authors | n | Aim | IM | C | Segmentation | Readers (n) |
FE | M | Main results | V |
|---|---|---|---|---|---|---|---|---|---|---|
| Aker et al. 2019 | 114 | Predict pCR in LARC | MRI | 1 | Manual ROI (T2WI) Rectal tu Post-CRT |
1(114) 2(20) |
FO TF |
N | ICC: 0.96 (intra-reader) ICC: 0.92 (inter-redear) Some FO TF correlate with pCR (AUC: 0.77-0.88) |
N |
| Horvat et al. 2018 | 114 | Predict pCR and compare it with subjective analysis in LARC | MRI (1.5T and 3T) | 1 | Manual ROI (T2WI) Rectal tu area Post-CRT |
2 consensus |
FO TF FO TF SO TF |
Y | Radiomics: AUC: 0.93, S: 1.00, Sp:0.91 T2WI: S: 0.57, Sp: 0.73 DWI: S: 0.63, Sp: 0.63 No difference between 1.5T and 3T |
N |
| Cusumano et al. 2018 | 198 | Predict pCR in LARC | MRI | 2 | Manual VOI (T2WI) Rectal tu Pre-treatment |
1 | Morf FO TF SO TF SupO |
Y | AUC: 0.77 (T) AUC: 0.79 (V) |
E |
| Cui et al. 2018 | 186 | Predict pCR in LARC | MRI | 1 | Manual ROI (T2WI, T1WI, ADC map) Pre-treatment |
1 (186) 2 (30) |
Morf FO TF SO TF |
Y | AUC radiomics (T): 0.94 AUC radiomics (V): 0.84 AUC nomogram* (T): 0.95 AUC nomogram* (V): 0.97 *radiomics + t-stage on MRI |
I |
| Bibault et al. 2018 | 95 | Predict pCR in LARC | CECT | 3 | Manual VOI Rectal tu Pre-treatment |
2 | Morf FO TF SO TF SupO |
Y | Deep neural network predicted pCR AUC: 0.72 | N |
| Liu et al. 2017 | 222 | Predict pCR in LARC Q | MRI (3T) | 1 | Manual VOI (T2WI and DWI) Rectal tu Pre- and post-CRT |
1 (222) 2 (80) |
FO TF SO TF SupO |
Y | Satisfactory inter- and intrareader AUC (T): 0.97 AUC (V): 0.98 C-index* (T): 0.98 C-index* (V): 0.97 *Radiomics + tu length post-CRT on MRI |
I |
| Chee et al. 2017 | 95 | Predict response to CRT and DFS | CECT | 1 | Manual ROI Rectal tu Pre-treatment |
2 consensus |
FO TF |
N | Several features were associated with response to CRT and some were associated with DFS | N |
| Meng et al. 2017 | 59 | Predict pCR response in LARC | MRI (3T) | 1 | Manual ROI (T2WI) Rectal tu Pre- and post-CRT |
2 | FO TF |
N | ICC (inter-reader): 0.60-0.99 Some features (kurtosis, energy, and entropy)predicted pCR with AUC varying from 0.67-0.76 |
N |
| Nie et al. 2016 | 48 | Predict pCR | MRI (3T) | Manual ROI (DCE and transferred to the other sequences) Rectal tu Pre- and post-CRT |
1 | FO TF SO TF |
Y | AUC: 0.85 (using mean ADC from DWI and one SO TF from DCE) Features from T1WI and T2WI showed lower prediction values |
I | |
| De Cecco et al. 2016 | 12 | Predict pCR using DWI, DCE, and radiomics | MRI (3T) | 1 | Manual ROI (T2WI) Rectal tu Pre- and post-CRT |
1 | FO TF |
N | Pre-treatment kurtosis better predicted pCR - AUC: 0.86 Radiomics better than DWI and DCE |
N |
| De Cecco et al. 2015 | 15 | Predict pCR in LARC | MRI (3T) | 1 | Manual ROI (T2WI) Rectal tu Pre- and post-CRT |
1 | FO TF |
N | Pre-treatment kurtosis AUC: 0.91 (cutoff <0.91) |
N |
ADC: apparent diffusion coefficients; AUC: area under the curve; C: centers; CECT: contrast enhanced computed tomography; CRT: chemoradiotherapy; DCE: dynamic contrast-enhanced; DFS: disease-free survival; DWI: diffusion-weighted imaging; E: external; FE: feature extraction; FO: first order; I: internal; ICC: intraclass correlation; IM: imaging modality; LARC: locally-advanced rectal cancer; M: model; Morf: morphological; N: no; pCR: pathological complete response; ROI: region of interest; S: sensitivity; SO: second order; Sp: specificity; SupO: superior order; T: training; TF: texture features; Tu: tumors; V: validation; VOI: volume of interest; WI: weighted images; Y: yes.
Table 3.
Characteristics of studies that have evaluated radiomics in predicting survival and presence of synchronous metastases in colorectal cancer. All studies were single-center studies.
| Author | n | Aim | IM | Segmentation | Reauers (n) |
FE | M | Main results | V |
|---|---|---|---|---|---|---|---|---|---|
| Van Helde et al. 2018 | 99 | Predict response and survival in patients with mCRC |
18F-FDG PET/CT |
Semiautomatic VOI All tu lesions Pre-treatment |
1 | Morf FO TF |
N | Some radiomics features correlates with survival | N |
| Liu et al. 2018 | 177 | Predict synchronous metastases in patients with rectal cancer | MRI (3T) | Manual VOI (T2WI) Rectal tu Pre-treatment |
2 | Morf FO TF SO TF |
Y | ICC> 0.80 AUC clinical (T): 0.79 AUC clinical (V): 0.77 AUC clinical-radiomics (T): 0.85 AUC clinical-radiomics (V): 0.83 |
I |
| Meng et al. 2018 | 108 | Predict DFS | MRI (3T) | Manual VOI (DCE) Rectal tu Pre-treatment |
2 consensus |
Morf FO TF SO TF SupO |
Y | ICC>0.80 C-index radiomics (T): 0.83 C-index clinical (T): 0.66 C-index both (T): 0.80 C-index radiomics (V): 0.77 C-index clinical (V): 0.64 C-index both (V): 0.79 |
I |
| Lovinfosse et al. 2017 | 86 | Predict survival in patients with LARC |
18F-FDG PET/CT |
Semiautomatic VOI Rectal tu Pre-treatment |
1 | FO TF SO TF SupO |
N | Coarseness was associated with DFS and DSS | N |
| Jalil et al. 2016 | 56 | Predict long term survival in patients treated with CRT | MRI (15T) | Manual ROI (T2WI) Rectal tu Pre- and post-CRT |
1 | FO TF |
N | Several texture features were associated with long term survival, particularly mean of positive pixels pre-CRT and kurtosis post-CRT | N |
ADC: apparent diffusion coefficients; DCE: dynamic contrast-enhanced; DFS: disease-free survival; DSS: disease-specific survival; FE: feature extraction; FO: first order; I: internal; ICC: intraclass correlation; IM: imaging modality; LARC: locally-advanced rectal cancer; M: model; Morf: morphological; N: no; ROI: region of interest; SO: second order; SupO: superior order; T: training; TF: texture features; Tu: tumors; V: validation; VOI: volume of interest; WI: weighted images; Y: yes; 18F-FDG PET: 18fluorodeoxyglucose positron emission tomography
Consistency in the attainment of quantitative features is one of the most important characteristics of a robust radiomics model. Therefore, more data regarding repeatability are necessary to enhance the accuracy of radiomic models. This is particularly relevant for CRC, since the delimitation of the tumor and normal bowel wall can be challenging, particularly after neoadjuvant CRT.
ii. Prediction of pathological complete response after CRT in patients with LARC
Currently, the main clinical challenge in rectal cancer is to preoperatively diagnose pathological complete response in patients with LARC after neoadjuvant CRT. As approximately 25% of the patients will demonstrate complete response after CRT, the concept of nonoperative approach has emerged to be viable (19) and has been solidified (20) as an alternative to surgery. However, at present there is no reliable method to diagnose complete response. In this scenario, radiomics has emerged as a promising tool that may serve as an imaging biomarker for tumor response. Table 1 summarizes the main studies that have evaluated this issue.
Studies have demonstrated that several first-order radiomics features extracted from T2 weighted images (WI) were associated with pathological complete response with area under the curve (AUC) values ranging from 0.67 to 0.91 (21-24). However, none of these studies created an advanced prediction model. Other authors have evaluated more complex radiomic features and created prediction models that demonstrated promising results (25-29). The AUC of these models in predicting pathological complete response ranged from 0.72 to 0.93 (25-30).
T2WI has been the main sequence used in the segmentation of the ROI or VOI of the tumor; however, a few studies have used other sequences including T1WI, diffusion-weighted imaging (DWI) and apparent diffusion coefficient (ADC) maps (27, 29). Cui et al. developed a nomogram incorporating T2WI, contrast-enhanced T1WI and ADC maps as well as imaging findings on MRI (T-stage during primary staging MRI) the nomogram achieved a C-index (concordance index) higher than 0.95 in both the training and validation set (27). Liu et al. developed a nomogram incorporating radiomics and tumor length on restaging MRI that showed an AUC of 0.98 (29).
Horvat et al. compared radiomics with conventional imaging interpretation and showed that the radiomics signature outperformed qualitative subjective analysis on diffusion weighted images and T2WI (25). Radiomics has also been shown to outperform quantitative assessment obtained on DWI and DCE (22).
While the majority of radiomic studies in patients with rectal cancer have used MRI-based features, two groups have exploited textural features extracted from computed tomography (CT) images. Chee et al. demonstrated that first order radiomics features from CT were associated with response to CRT (32), and Bibault et al. reported that a prediction model developed using deep learning had an AUC of 0.72 (28).
Despite the encouraging results, there is still a long way to go before radiomics can be implemented in clinical practice. The methodologies in these studies differ: some have involved mathematical corrections that may have resulted in overoptimistic results; a few performed internal validation; and only one performed external validation. Consequently, further studies, especially with large datasets as well as consistent methodology and validation, are needed to assess the potential of radiomics in the prediction of pathological complete response.
iii. Prediction of other pathological features
Besides the prediction of pathologic complete response to neoadjuvant CRT, studies have evaluated radiomics for the prediction of other pathological features based on the imaging features obtained during primary staging (Table 2). Some radiomic features extracted from CT (33) and MRI using T2WI (34) and ADC map (31) have shown promising results in the prediction of T-staging.
Table 2.
Characteristics of studies that have evaluated radiomics in predicting several pathological features after surgery in colorectal cancer. All studies were single-center studies.
| Author | n | Aim | IM | Segmentation | Readers (n) |
FE | M | Main results | V |
|---|---|---|---|---|---|---|---|---|---|
| Li et al. 2018 | 50 | Predict pathological features in rectal tu | MRI (3T) | Manual VOI (ADC map) Rectal tu and normal wall Pre-treatment |
2 | FO TF SO TF |
N c | ICC: 0.72-0.97 (intra and inter-reader) Entropy predicts T stage |
N |
| Sun et al. 2018 | 119 | Predict pathological features in rectal tu | MRI (3T) | Manual VOI (T2WI) Rectal tu Pre-treatment |
2 or 3 consensus |
Morf FO TF SO TF SupO |
Y | Better performance in predict T-stage AUC: 0.85 |
I |
| Huang et al. 2018 | 346 | Predict perineural invasion in CRC | CECT | Manual ROI CRC tu Pre-treatment |
1 (346) 2 (80) |
FO TF SO TF |
Y | ICC: 0.75-0.90 (inter-reader) ICC: 0.79-0.91 (intra-reader) C-index radiomics (T): 0.81 C-index radiomics (V): 0.78 C-index radiomics + CEA (T): 0.81 C-index radiomics + CEA (V): 0.80 |
I |
| Huang et al. 2018 | 366 | Differentiate high-grade from low-grade CRC | CECT | Manual VOI CRC tu Pre-treatment |
2 | FO TF SO TF |
Y | ICC> 0.75 AUC (T): 0.81 AUC (V): 0.74 AUC rectal tu: 0.89 AUC colon tu: 0.72 |
I |
| Chen et al. 2018 | 115 | Predict lymph node metastasis in rectal tu | ERU CECT SWE | Manual ROI and VOI Rectal tu Pre-treatment |
2 | Morf FO TF SO TF |
Y | C-index radiomics (T): 0.74 C-index size criteria ERUS (T): 0.55 C-index size criteria CT (T): 0.74 C-index radiomics + size (T): 0.87 C-index radiomics (V): 0.70 C-index size criteria ERUS (V):0.54 C-index size criteria CT (V): 0.67 C-index radiomics + size (V): 0.86 |
I |
| Liang et al. 2016 | 494 | Differentiate stages I-II vs III-IV CRC | CECT | Manual ROI CRC tu Pre-treatment |
1 | FO TF |
Y | AUC clinical (T): 0.63 AUC radiomics (T): 0.79 AUC both (T): 0.81 AUC clinical (V): 0.59 AUC radiomics (V): 0.71 AUC both (V): 0.72 |
I |
| Liu et al. 2016 | 60 | Predict T (pT1-2 vs pT3-4) and N stages (pN0 vs pN1-N2) in rectal tu | MRI (3T) | Manual VOI (ADC map) Rectal tu Pre-treatment |
2 | FO TF SO TF |
N | ICC: 0.78-0.97 Skewness and entropy: T stage (AUC: 0.74) ADC max and entropy: N stage (AUC: 0.75) |
N |
| Huang et al. 2016 | 326 | Predict node metastasis in CRC | CECT | Manual ROI (CECT) Colorectal tu Pre-treatment |
2 | FO TF SO TF |
Y | ICC: 0.76-0.91 (inter-reader) ICC: 0.81-0.95 (intra-reader) Nomogram with radiomics, CT node status and CEA: C-index: 0.74 (T) / C-index: 0.78 (V) |
I |
ADC: apparent diffusion coefficients; AUC: area under the curve; CEA: carcinoembryonic antigen; CECT: contrast enhanced computed tomography; CRT: chemoradiotherapy; DCE: dynamic contrast-enhanced; DFS: disease-free survival; E: external; ERUS: endorectal ultrasound; FE: feature extraction; FO: first order; I: internal; ICC: intraclass correlation; IM: imaging modality; LARC: locally-advanced rectal cancer; M: model; Morf: morphological; N: no; pCR: pathological complete response; ROI: region of interest; S: sensitivity; SO: second order; Sp: specificity; SupO: superior order; shear-wave elastography; T: training; TF: texture features; Tu: tumors; V: validation; VOI: volume of interest; WI: weighted images; Y: yes.
Radiomics has also been studied in the prediction of lymph node metastasis in patients with CRC using CT (35) and in patients with rectal cancer using endorectal ultrasound, CT and shear-wave elastography (36). In the first study, Huang et al. developed a nomogram using radiomic features, nodal status on qualitative CT and carcinoembryonic antigen (CEA) positivity; this nomogram achieved a C-index of 0.74 in training set and 0.78 in validation set (35). In the second study, Chen et al. produced several nomograms; the multiparametric nomogram obtained the higher performance (c-index of 0.87 in the training section and 0.86 in the validation section) compared with the conventional nomogram that was based on enhancement changes in the tumor (36).
Two studies have assessed radiomics on CT to predict perineural invasion (37) and to differentiate high- from low-grade CRC (38). In the first study, nomogram incorporating a radiomics signature and CEA level which achieved a c-index of 0.82 (37). In the second study, radiomics had a moderate AUC (0.74-0.81) for differentiating high- from low-histological grade CRC,; however, the performance among rectal tumors was higher than in colon tumors (AUC: 0.89 vs. 0.72) (38).
iv. Prediction of survival
Radiomics analysis based on 18F-FDG PET/CT (39, 40) and MRI (41,42) has also been used to estimate survival parameters in patients with CRC (Table 3). Several radiomic features were found to correlate with survival and overall (39, 40, 42). Meng et al. found that the model incorporating both radiomics and clinical data obtained the best performance (41). Radiomics also was associated with the presence of synchronous metastasis in patients with rectal cancer, with AUC above 0.80 for the models that associated clinical variables (43).
3. Radiogenomics
As the interest in genetic testing has grown over the last several decades, the need to match diagnostic imaging to gene expression patterns of different cancers has emerged as a critical area of research. In radiogenomics, qualitative and/or quantitative imaging features are extracted and correlated with genetic profiles of the imaged tissue (8). The development of high quality, reproducible radiogenomics profiling tools has tremendous potential to further the role of imaging biomarkers, particularly in oncologic imaging.
To date, there is relatively limited published literature on radiogenomics in CRC. One area of radiogenomics in CRC that has gained traction is the KRAS mutation status for a given patient. KRAS mutations in CRC are present in approximately 40% of cases (49) and are associated with low responsiveness to drugs targeting epidermal growth factor receptors (EGFR). Currently, the published literature on radiogenomic features of KRAS status in CRC has focused on both the primary neoplasm and distant metastases (49-56). It is worth mentioning that high concordance, on the order of 96% and 97%, has been reported for the KRAS status of primary tumors and metastases in a given patient (57, 58), which implies that when a CRC metastasis has been biopsied and KRAS status has been determined, it is almost always the case that the primary tumor will have the same KRAS mutation status.
In a cohort of patients with rectal cancer being evaluated with MRI, Shin et al. (49) found that tumors with KRAS mutations are associated with polypoid morphology, increased axial length, increased axial-to-longitudinal tumor ratio and N2 nodal status. An additional study by Lubner et al. (59) involved studying the radiomic features of hepatic metastases in CRC using texture features and included a radiogenomic analysis to identify associations of the radiomic features with KRAS mutations. They found that the skewness feature was inversely associated with the presence of a KRAS mutation.
Beyond CT and MRI, a number of studies have examined the role of 18F-FDG PET in the radiogenomics assessment of KRAS mutations in CRC. Mao et al. found that CRC liver metastases had a higher SUVmax when KRAS mutations were present on both early and delayed acquisitions (50). Kawada et al. limited their analysis to only tumors greater than 10 mm in size and reported that KRAS mutation can be reliably predicted with an accuracy of 71.4% using an SUVmax cutoff of greater than 6.0 in their cohort (53). Lastly, Chen et al. reported in two studies that higher SUVmax and TW40% were associated with KRAS mutated tumors (56, 60). They also reported an association with these parameters in TP53 mutated tumors in one of those studies (60). However, a number of studies have refuted these findings and found that PET parameters could not predict KRAS status in a meaningful way (52, 55, 61). In summary, the role of 18F-FDG PET in radiogenomics for the assessment of KRAS status in CRC remains controversial but may still have a great potential.
The potential for future research in radiogenomics analysis of CRC is tremendous. The ability to determine the presence or absence of KRAS mutations in CRC reliably with routine diagnostic imaging would be clinically useful. It is interesting that to date, most of the research on this question has focused on 18F-FDG PET in patients with CRC; however, this modality is less commonly used for staging in routine clinical practice. There may be more practical potential to investigate these questions using CT and MRI, which are performed in the vast majority of patients with newly diagnosed CRC in the United States today. The opportunity to use radiogenomics to study KRAS status as well as other mutations in CRC is wide open.
4. Conclusion
Radiomics and radiogenomics may change the practice of medicine, particularly for patients with CRC. Thus far, several studies have evaluated the use of radiomics for this patient population; however, there are challenges to be overcome before its routine implementation including challenges related to sample size, model design and interpretability, and the lack of robust multicenter validation set. Radiologists should be aware of the basic concepts, benefits, pitfalls and limitations of new radiomic and radiogenomic techniques to achieve a balanced interpretation of the results.
Acknowledgments
Funding: This work was supported in part through the NIH/NCI Cancer Center Support
Grant P30 CA008748.
Footnotes
Conflicts of interest: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278(2):563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu W, Parmar C, Grossmann P, et al. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front Oncol. 2016;6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beig N, Khorrami M, Alilou M, et al. Perinodular and Intranodular Radiomic Features on Lung CT Images Distinguish Adenocarcinomas from Granulomas. Radiology. 2018:180910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kickingereder P, Gotz M, Muschelli J, et al. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clin Cancer Res. 2016;22(23):5765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119(3):480–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horvat N, Veeraraghavan H, Khan M, et al. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology. 2018;287(3):833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreassen CN, Alsner J, Overgaard J. Does variability in normal tissue reactions after radiotherapy have a genetic basis--where and how to look for it? Radiother Oncol. 2002;64(2):131–40. [DOI] [PubMed] [Google Scholar]
- 8.Pinker K, Shitano F, Sala E, et al. Background, current role, and potential applications of radiogenomics. J Magn Reson Imaging. 2018;47(3):604–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sala E, Mema E, Himoto Y, et al. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol. 2017;72(1):3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganeshan B, Miles KA. Quantifying tumour heterogeneity with CT. Cancer Imaging. 2013;13:140–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haralick RM, Shanmugam K, Dinstein I. Textural Features for Image Classification. Ieee Transactions on Systems Man and Cybernetics. 1973;Smc3(6):610–21. [Google Scholar]
- 12.Larue RT, Defraene G, De Ruysscher D, et al. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol. 2017;90(1070):20160665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, et al. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology. 2018;288(2):407–15. [DOI] [PubMed] [Google Scholar]
- 14.Lubner MG, Smith AD, Sandrasegaran K, et al. CT Texture Analysis: Definitions, Applications, Biologic Correlates, and Challenges. Radiographics. 2017;37(5):1483–503. [DOI] [PubMed] [Google Scholar]
- 15.Lubner MG, Stabo N, Abel EJ, et al. CT Textural Analysis of Large Primary Renal Cell Carcinomas: Pretreatment Tumor Heterogeneity Correlates With Histologic Findings and Clinical Outcomes. AJR Am J Roentgenol. 2016;207(1):96–105. [DOI] [PubMed] [Google Scholar]
- 16.Miles KA, Ganeshan B, Hayball MP. CT texture analysis using the filtration-histogram method: what do the measurements mean? Cancer Imaging. 2013;13(3):400–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: Synthetic minority over-sampling technique. Journal of Artificial Intelligence Research. 2002;16:321–57. [Google Scholar]
- 18.Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A. 2015;112(46):E6265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habr-Gama A, Perez RO, Nadalin W, et al. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: long-term results. Ann Surg. 2004;240(4):711–7; discussion 7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith JJ, Strombom P, Chow OS, et al. Assessment of a Watch-and-Wait Strategy for Rectal Cancer in Patients With a Complete Response After Neoadjuvant Therapy. JAMA Oncol. 2019:e185896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Cecco CN, Ganeshan B, Ciolina M, et al. Texture analysis as imaging biomarker of tumoral response to neoadjuvant chemoradiotherapy in rectal cancer patients studied with 3-T magnetic resonance. Invest Radiol. 2015;50(4):239–45. [DOI] [PubMed] [Google Scholar]
- 22.De Cecco CN, Ciolina M, Caruso D, et al. Performance of diffusion-weighted imaging, perfusion imaging, and texture analysis in predicting tumoral response to neoadjuvant chemoradiotherapy in rectal cancer patients studied with 3T MR: initial experience. Abdom Radiol (NY). 2016;41(9):1728–35. [DOI] [PubMed] [Google Scholar]
- 23.Meng Y, Zhang C, Zou S, et al. MRI texture analysis in predicting treatment response to neoadjuvant chemoradiotherapy in rectal cancer. Oncotarget. 2018;9(15):11999–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aker M, Ganeshan B, Afaq A, et al. Magnetic Resonance Texture Analysis in Identifying Complete Pathological Response to Neoadjuvant Treatment in Locally Advanced Rectal Cancer. Dis Colon Rectum. 2019;62(2):163–70. [DOI] [PubMed] [Google Scholar]
- 25.Horvat N, Veeraraghavan H, Khan M, et al. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology. 2018:172300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cusumano D, Dinapoli N, Boldrini L, et al. Fractal-based radiomic approach to predict complete pathological response after chemo-radiotherapy in rectal cancer. Radiol Med. 2017. [DOI] [PubMed] [Google Scholar]
- 27.Cui Y, Yang X, Shi Z, et al. Radiomics analysis of multiparametric MRI for prediction of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Eur Radiol. 2018. [DOI] [PubMed] [Google Scholar]
- 28.Bibault JE, Giraud P, Housset M, et al. Deep Learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. 2018;8(1):12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Zhang XY, Shi YJ, et al. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin Cancer Res. 2017;23(23):7253–62. [DOI] [PubMed] [Google Scholar]
- 30.Nie K, Shi L, Chen Q, et al. Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. Clin Cancer Res. 2016;22(21):5256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Jiang Z, Guan Y, et al. Whole-lesion Apparent Diffusion Coefficient First- and Second-Order Texture Features for the Characterization of Rectal Cancer Pathological Factors. J Comput Assist Tomogr. 2018;42(4):642–7. [DOI] [PubMed] [Google Scholar]
- 32.Chee CG, Kim YH, Lee KH, et al. CT texture analysis in patients with locally advanced rectal cancer treated with neoadjuvant chemoradiotherapy: A potential imaging biomarker for treatment response and prognosis. PLoS One. 2017;12(8):e0182883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang C, Huang Y, He L, et al. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget. 2016;7(21):31401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Hu P, Wang J, et al. Radiomic features of pretreatment MRI could identify T stage in patients with rectal cancer: Preliminary findings. J Magn Reson Imaging. 2018. [DOI] [PubMed] [Google Scholar]
- 35.Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol. 2016;34(18):2157–64. [DOI] [PubMed] [Google Scholar]
- 36.Chen LD, Liang JY, Wu H, et al. Multiparametric radiomics improve prediction of lymph node metastasis of rectal cancer compared with conventional radiomics. Life Sci. 2018;208:55–63. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y, He L, Dong D, et al. Individualized prediction of perineural invasion in colorectal cancer: development and validation of a radiomics prediction model. Chin J Cancer Res. 2018;30(1):40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X, Cheng Z, Huang Y, et al. CT-based Radiomics Signature to Discriminate High-grade From Low-grade Colorectal Adenocarcinoma. Acad Radiol. 2018;25(10):1285–97. [DOI] [PubMed] [Google Scholar]
- 39.van Helden EJ, Vacher YJL, van Wieringen WN, et al. Radiomics analysis of pre-treatment [Eur J Nucl Med Mol Imaging. 2018;45(13):2307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lovinfosse P, Polus M, Van Daele D, et al. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur J Nucl Med Mol Imaging. 2018;45(3):365–75. [DOI] [PubMed] [Google Scholar]
- 41.Meng Y, Zhang Y, Dong D, et al. Novel radiomic signature as a prognostic biomarker for locally advanced rectal cancer. J Magn Reson Imaging. 2018. [DOI] [PubMed] [Google Scholar]
- 42.Jalil O, Afaq A, Ganeshan B, et al. Magnetic resonance based texture parameters as potential imaging biomarkers for predicting long-term survival in locally advanced rectal cancer treated by chemoradiotherapy. Colorectal Dis. 2017;19(4):349–62. [DOI] [PubMed] [Google Scholar]
- 43.Liu H, Zhang C, Wang L, et al. MRI radiomics analysis for predicting preoperative synchronous distant metastasis in patients with rectal cancer. Eur Radiol. 2018. [DOI] [PubMed] [Google Scholar]
- 44.Aker M, Boone D, Chandramohan A, et al. Diagnostic accuracy of MRI in assessing tumor regression and identifying complete response in patients with locally advanced rectal cancer after neoadjuvant treatment. Abdom Radiol (NY). 2018. [DOI] [PubMed] [Google Scholar]
- 45.Yang L, Dong D, Fang M, et al. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol. 2018;28(5):2058–67. [DOI] [PubMed] [Google Scholar]
- 46.Meng X, Xia W, Xie P, et al. Preoperative radiomic signature based on multiparametric magnetic resonance imaging for noninvasive evaluation of biological characteristics in rectal cancer. Eur Radiol. 2018. [DOI] [PubMed] [Google Scholar]
- 47.Gourtsoyianni S, Doumou G, Prezzi D, et al. Primary Rectal Cancer: Repeatability of Global and Local-Regional MR Imaging Texture Features. Radiology. 2017;284(2):552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Badic B, Desseroit MC, Hatt M, Visvikis D. Potential Complementary Value of Noncontrast and Contrast Enhanced CT Radiomics in Colorectal Cancers. Acad Radiol. 2018 [DOI] [PubMed] [Google Scholar]
- 49.Shin YR, Kim KA, Im S, et al. Prediction of KRAS Mutation in Rectal Cancer Using MRI. Anticancer Res. 2016;36(9):4799–804. [DOI] [PubMed] [Google Scholar]
- 50.Mao W, Zhou J, Zhang H, et al. Relationship between KRAS mutations and dual time point (18)F-FDG PET/CT imaging in colorectal liver metastases. Abdom Radiol (NY). 2018. [DOI] [PubMed] [Google Scholar]
- 51.Pershad Y, Govindan S, Hara AK, et al. Using Naive Bayesian Analysis to Determine Imaging Characteristics of KRAS Mutations in Metastatic Colon Cancer. Diagnostics (Basel). 2017;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oner AO, Budak ES, Yildirim S, et al. The value of (18)FDG PET/CT parameters, hematological parameters and tumor markers in predicting KRAS oncogene mutation in colorectal cancer. Hell J Nucl Med. 2017;20(2):160–5. [DOI] [PubMed] [Google Scholar]
- 53.Kawada K, Toda K, Nakamoto Y, et al. Relationship Between 18F-FDG PET/CT Scans and KRAS Mutations in Metastatic Colorectal Cancer. J Nucl Med. 2015;56(9):1322–7. [DOI] [PubMed] [Google Scholar]
- 54.Miles KA, Ganeshan B, Rodriguez-Justo M, et al. Multifunctional imaging signature for V-KI-RAS2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations in colorectal cancer. J Nucl Med. 2014;55(3):386–91. [DOI] [PubMed] [Google Scholar]
- 55.Krikelis D, Skoura E, Kotoula V, et al. Lack of association between KRAS mutations and 18F-FDG PET/CT in Caucasian metastatic colorectal cancer patients. Anticancer Res. 2014;34(5):2571–9. [PubMed] [Google Scholar]
- 56.Chen SW, Chiang HC, Chen WT, et al. Correlation between PET/CT parameters and KRAS expression in colorectal cancer. Clin Nucl Med. 2014;39(8):685–9. [DOI] [PubMed] [Google Scholar]
- 57.Mariani P, Lae M, Degeorges A, et al. Concordant analysis of KRAS status in primary colon carcinoma and matched metastasis. Anticancer Res. 2010;30(10):4229–35. [PubMed] [Google Scholar]
- 58.Santini D, Loupakis F, Vincenzi B, et al. High concordance of KRAS status between primary colorectal tumors and related metastatic sites: implications for clinical practice. Oncologist. 2008;13(12):1270–5. [DOI] [PubMed] [Google Scholar]
- 59.Lubner MG, Stabo N, Lubner SJ, et al. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40(7):2331–7. [DOI] [PubMed] [Google Scholar]
- 60.Chen SW, Lin CY, Ho CM, et al. Genetic Alterations in Colorectal Cancer Have Different Patterns on 18F-FDG PET/CT. Clin Nucl Med. 2015;40(8):621–6. [DOI] [PubMed] [Google Scholar]
- 61.Lovinfosse P, Koopmansch B, Lambert F, et al. (18)F-FDG PET/CT imaging in rectal cancer: relationship with the RAS mutational status. Br J Radiol. 2016;89(1063):20160212. [DOI] [PMC free article] [PubMed] [Google Scholar]