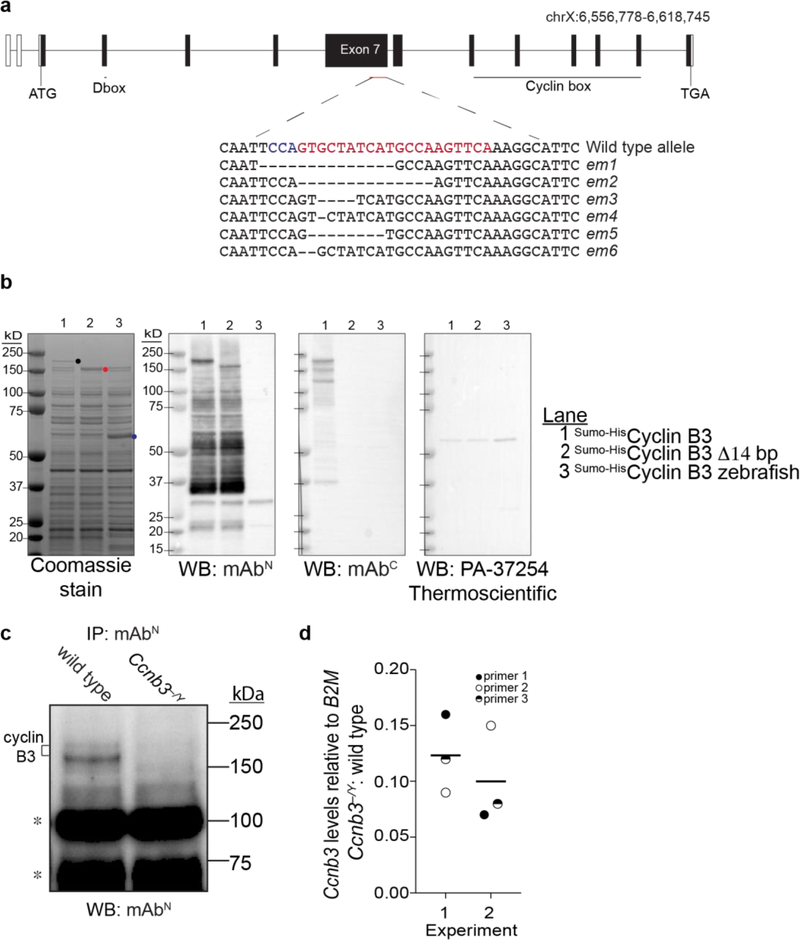
Figure 2: CRISPR/Cas9-mediated Ccnb3 allele generation and confirmation.
a) Ccnb3 gene diagram. Open boxes show 5’ and 3’ untranslated regions and filled boxes show the coding regions. The CRISPR-Cas9 targeted position is indicated with protospacer adjacent motif (PAM) in blue and single guide RNA sequence in red. Sequences are shown for six em alleles recovered. The em1 allele was characterized in this study and Karasu et al. 2019.
b) Expression and detection of recombinant cyclin B3 proteins. Constructs contained either full-length coding sequence or the 14-bp deletion of the em1 allele fused to the Sumo-His6 tag (Wasmuth and Lima 2012). Zebrafish cyclin B3 was used as a negative control for detection by WB. Proteins were expressed in E. coli and equal aliquots of soluble extracts were fractionated on SDS-PAGE and stained with Coomassie blue or analyzed by WB with the indicated antibody. Dots indicate positions of the largest detected proteins encoded by each construct. Lower molecular weight bands detected by WB with mAbN or mAbC presumably are proteolytic fragments and/or products of premature translation termination.
c) IP-WB for cyclin B3. Whole testis extracts from Ccnb3−/Y and wild-type adults were analyzed using mAbN Note that IP-WB using only this antibody is less sensitive and gives higher background than when combining mAbN with mAbC (Fig 1b). Asterisks indicate lower molecular weight bands independent of genotype, possibly from immunoglobulin and other contaminants in the IP elute.
d) RT-qPCR analysis of whole testis RNA extracted from Ccnb3−/Y and wild-type adults. The plotted values represent the fold difference between mutant and wild type for Ccnb3 levels normalized to B2M. Three different pairs of Ccnb3 primers were used. Two independent experiments were performed, each containing a separate mutant–wild type pair. Lines indicate means for each experiment.