Fig. 1.
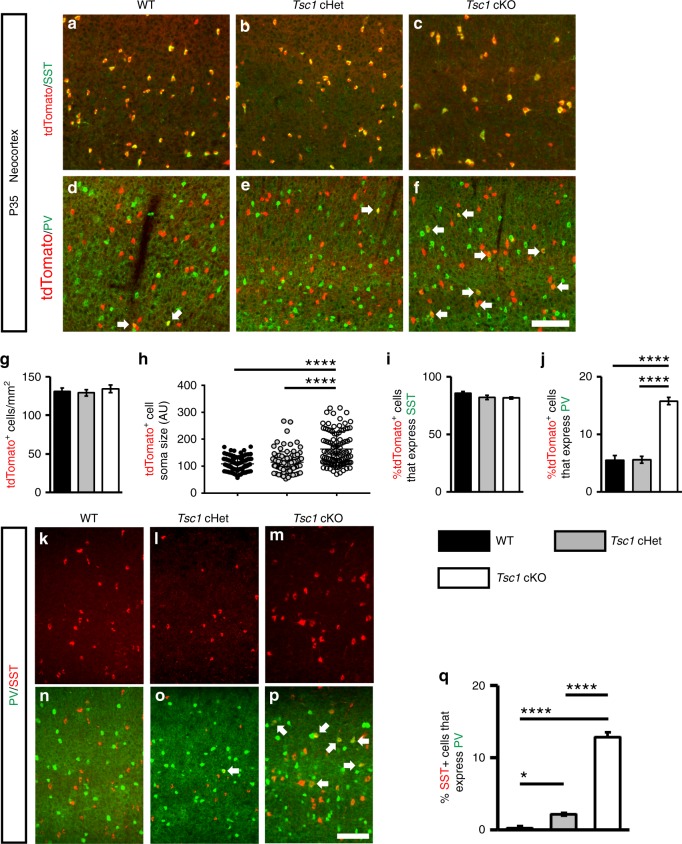
Post-mitotic deletion of Tsc1 in SST-Cre-lineages results in increased PV expression. Coronal immunofluorescent images from WT, Tsc1 cHets or Tsc1 cKOs P35 neocortices, showing co-localization of tdTomato (SST-Cre-lineages) with either SST (a–c) or PV (d–f). Arrows d–f denote cells in the SST-Cre-lineage that express PV. g Quantification of the number of tdTomato+ cells/mm2 (cell density) in the neocortex: n = 6 (WT), 6 (cHet) and 4 (cKO) mice, One-way ANOVA (F2, 13 = 0.35, P = 0.7). h Quantification of tdTomato+ cell soma size: n = 4 mice per genotype (100 cells measured from each genotype), (F2, 297 = 44.9, P < 0.0001; cKO vs. WT and cHet, p < 0.0001). (AU) arbitrary units. g, h Quantification of the %tdTomato+ cells that co-express either SST (i) or PV (j): Chi-squared test with Yate’s correction, (p < 0.0001, all groups); n = 6 WT (1268 cells), n = 6 cHet (1371 cells) and n = 4 cKO (1218 cells) for SST; n = 5 WT (1281 cells), n = 6 cHet (1600 cells) and n = 4 cKO (1220 cells) for PV. k–p Immunofluorescent images showing SST and PV co-labeling in the neocortex. q Quantification of the %SST+ CINs that co-express PV: Chi-squared test with Yate’s correction, (WT vs. cHet, p = 0.03, cKO vs. WT or cHet, p < 0.0001), n = 3 mice, all groups. Data are represented as mean ± SD in (h) and ± SEM in all other graphs. Scale bars in f, p = 100 µm. *p < 0.05, ****p < 0.0001. Source data are provided as a Source Data file