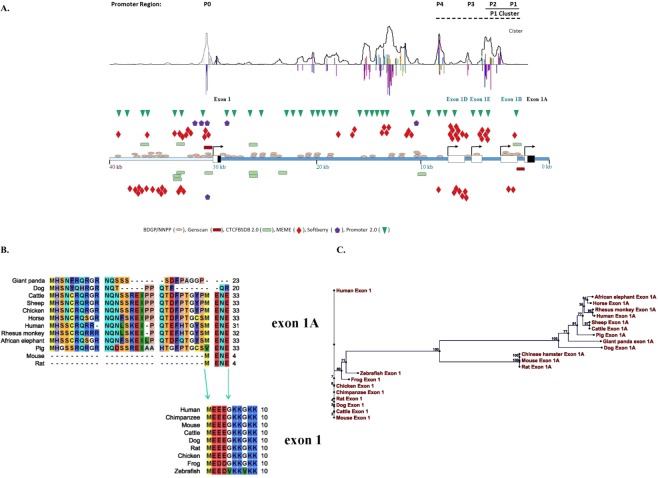
Figure 1.
Genomic structure, conservation, and variability among alterative start-codon exons and promotors of CCM2. (A) The complex promoter regions of human CCM2 locus were defined with bioinformatics (promoter prediction software from top to bottom: Cister, promotor2.0, Softberry, MEME, CTCFBSDB, BDGP/NNPP and Genscan as indicated by different colors). Symbols on top of DNA templates are on positive strand, below are on negative strand. The single promoter for the original bonafide start-codon exon, exon 1, simply lies immediately upstream of the transcription start site for exon 1, as P0. The promoter region for exon 1A is much more complicated. Although a seemingly weak promoter, P1 lies immediately upstream of its transcription-start site; in addition, there are three relatively strong promoters (P2-P4) upstream adjacent to P1 promoter. Three exons (exon 1B, 1D, 1E) with the transcription start site driven by these three promoters (P2-P4), respectively, usually skip exon 1A (coding exon), presumably to down-regulate the transcription level of group B CCM2 isoforms. Genomic structure of 5′ region of the human CCM2 locus is schematically summarized in this map. Noncoding region within a transcription-start exon labeled as white box while coding region within the exon labeled as black box. (B) Multiple-alignment between two alterative start codon exons (exon 1 and exon 1A) across species reveals a vertebrate-specific exon 1 and a mammalian-specific exon 1A and their evolutional relationship. Exon 1A is evolutionarily evolved from exon1 with its C-terminus homolog to the N-terminus of exon1. (C) Phylogenetic relationships between exon 1A and exon 1 among CCM2 isoforms across species based on neighbor joining (NJ) method which hypothesizes a stochastic process in different lineages during evolution.