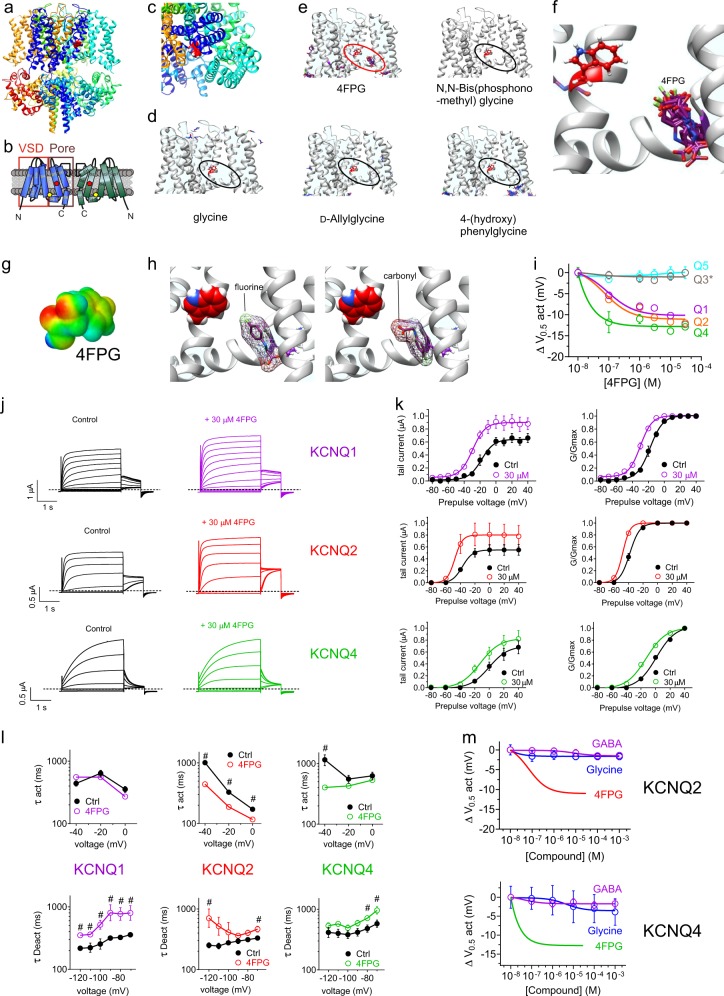
Fig. 2.
In silico prediction and in vitro validation of a KCNQ-activating glycine derivative. All error bars indicate SEM. a Chimeric KCNQ1/KCNQ3 structural model (red, KCNQ3-W265). b Topological representation of KCNQ channel showing two of the four subunits, without domain swapping for clarity. Pentagon, approximate position of KCNQ3-W265; VSD, voltage sensing domain. c Close-up extracellular view of KCNQ1/KCNQ3 structural model (red, KCNQ3-W265). d Docking result showing predicted lack of binding of glycine to the KCNQ1/KCNQ3 structural model. Red, KCNQ3-W265; black oval highlights lack of glycine binding in the typical binding zone for GABA and its metabolites and analogs. e Docking results for various glycine derivatives illustrated in Fig. 1 showing predicted binding of 4FPG within the GABA binding pocket (highlighted by red oval) but not of the other molecules (black ovals). All predicted binding configurations shown overlaid for each molecule. f Close-up of predicted binding poses of 4FPG within the GABA binding pocket. g Surface electrostatic potential plot of 4FPG. h Comparison of two different predicted orientations of 4FPG within the KCNQ binding pocket, as predicted by SwissDock. i 4FPG dose responses for homomeric KCNQ1, 2, 3*, 4, and 5 channels expressed in oocytes, quantified as shift in the voltage dependence of channel activation (ΔV0.5act); n = 4–6. j Mean traces showing effects of 4FPG (30 µM) on KCNQ1, KCNQ2 and KCNQ4; n = 4–6. k Effects of 4FPG (30 µM) on KCNQ1, KCNQ and KCN raw and normalized (G/Gmax) tail current, calculated from traces as in panel j; n = 4–6. l Effects of 4FPG (30 µM) on KCNQ1, KCNQ2, and KCNQ4 activation (act) and deactivation (Deact) rates, fitted as a single exponential function (τ); n = 4–6. m 4FPG dose responses for KCNQ2 and KCNQ4 compared to those of glycine and GABA, quantified as shift in voltage dependence of activation (ΔV0.5act); n = 5–6