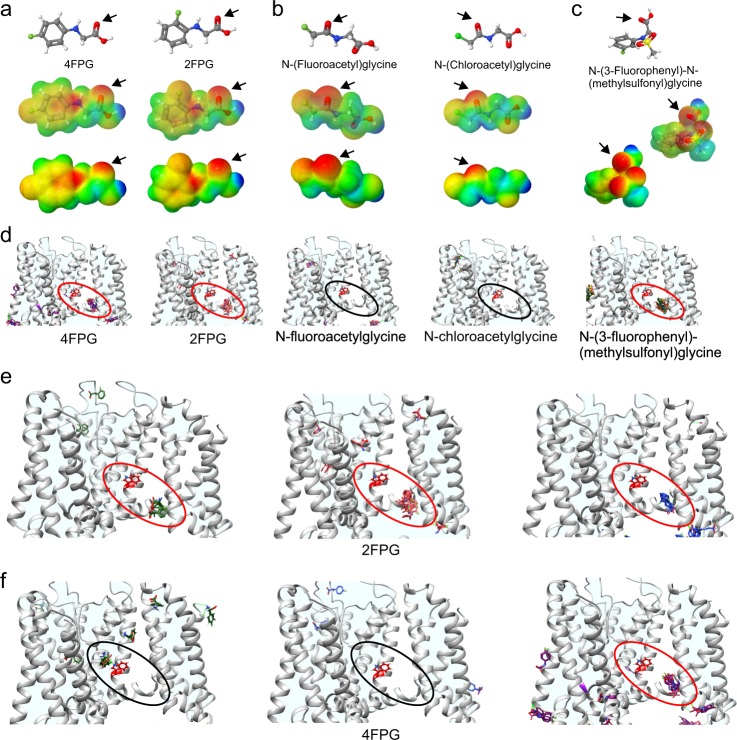
Fig. 3.
In silico prediction of 4FPG-related KCNQ-activating glycine derivatives. a Chemical properties of 4FPG versus 2FPG: structure, electrostatic surface potentials (red, electron-dense; blue, electron-poor; green, neutral) and an overlay of the two, all calculated and plotted using Jmol. Arrows, native glycine carbonyl. b, c Chemical properties of 4FPG-related glycine derivatives, parameters as in (a). Arrows, non-native glycine carbonyls for N-(fluoroacetyl)glycine and N-(Chloroacetyl)glycine; native glycine carbonyl for 3FMSG. d Docking results showing predicted binding (red ovals) or lack thereof (black ovals) of the compounds in (a–c) to the GABA binding pocket in the KCNQ1/KCNQ3 structural model. Red side-chain, KCNQ3-W265. e Docking results showing predicted binding (red ovals) of three different conformational forms of 2FPG to the GABA binding pocket in the KCNQ1/KCNQ3 structural model. Red side-chain, KCNQ3-W265. f Docking results showing predicted binding (red oval) or lack thereof (black ovals) of three different conformational forms of 4FPG to the GABA binding pocket in the KCNQ1/KCNQ3 structural model. Red side-chain, KCNQ3-W265