Abstract
Background
Vector-borne diseases are among the leading causes of morbidity and mortality in humans and animals. In the Afrotropical region, some are transmitted by Culicoides, such as Akabane, bluetongue, epizootic haemorrhagic fever and African horse sickness viruses. Bluetongue virus infection has an enormous impact on ruminant production, due to its high morbidity and mortality rates.
Methods
A nationwide Culicoides trapping campaign was organized at the end of the 2012 rainy season in Senegal. A Maximum Entropy approach (MaxEnt), Boosted Regression Tree (BRT) method and Ecological Niche Factor Analysis (ENFA) were used to develop a predictive spatial model for the distribution of Culicoides, using bio-climatic variables, livestock densities and altitude.
Results
The altitude, maximum temperature of the warmest month, precipitation of the warmest quarter, mean temperature of the wettest quarter, temperature seasonality, precipitation of the wettest quarter and livestock density were among the most important factors to predict suitable habitats of Culicoides. Culicoides occurrences were, in most of the cases, positively correlated to precipitation variables and livestock densities; and negatively correlated to the altitude and temperature indices. The Niayes area and the Groundnut basin were the most suitable habitats predicted.
Conclusion
We present ecological niche models for different Culicoides species, namely C. imicola, C. oxystoma, C. enderleini and C. miombo, potential vectors of bluetongue virus, on a nationwide scale in Senegal. Through our modelling approach, we were able to determine the effect of bioclimatic variables on Culicoides habitats and were able to generate maps for the occurrence of Culicoides species. This information will be helpful in developing risk maps for disease outbreaks.
Keywords: Vector-borne diseases, Afrotropical region, Culicoides, Bluetongue, Ecological modelling, MaxEnt, Boosted Regression Tree, Ecological Niche Factor Analysis, Suitable habitats
Background
Vector-borne diseases are among the leading causes of morbidity and mortality in humans and animals. In the Afrotropical region, Culicoides species are the main vectors for highly devastating viruses such as Akabane, bluetongue (BT), epizootic haemorrhagic fever and African horse sickness (AHS) [1, 2]. BT, AHS and EHD are listed among the reportable diseases of the World Organization for Animal Health (OIE). Bluetongue virus (BTV) is transmitted to hosts, both wild and domestic ruminants, by the bites of midges of the genus Culicoides, and infections can lead to the host’s death. Few studies exist on the epidemiological situation in Senegal. Two of them estimated sero-prevalence varying between 30 and 59% for cattle and sheep [3, 4]. Understanding the trophic behaviour and the spatial dynamics of Culicoides species could help in controlling the spread of BT.
Studies on the trophic behaviour of Culicoides species have shown that these midges feed on various hosts in Afrotropical region, but mainly on mammals and birds [5–7]. The frequency of blood meals is 3 to 5 days depending on the availability of hosts, which are necessary to complete their gonotrophic cycle [8, 9]. After egg maturation, which occurs 2 to 4 days after the blood meal [10], females seek oviposition sites to deposit their eggs. The number of eggs laid varies from 30 to 250. A vermiform larva is free of pseudopods within 3 to 10 days after being hatched [11]. Larvae of Culicoides species live in various habitats, but they are mostly wet and enriched in organic matter of animal or plant origin [12–17].
Adult ecology can be studied taking a purely statistical approach. Predictive modelling of species geographical distributions, based on environmental conditions, is a major technique in analytical biology, with applications in conservation and reserve planning, ecology, evolution, epidemiology, invasive-species management and other fields [18–21]. Sometimes both presence and absence data are available for developing models, in which case general-purpose statistical methods can be used [22, 23]. However, whilst presence data can be collected through trapping campaigns, absence data are rather more difficult to collect and interpret.
Species distribution models (SDM) can be used to predict the distribution of species. Several methods, belonging to different classes, can be used to evaluate SDM:”profile” such as Domain [24], Mahalanobis distance [25], regression such as Generalized Additive Models (GAM) [26, 27]; machine learning such as Random Forest [28], Support Vector Machines (SVM) [29], Boosted Regression Trees [30], MaxEnt [31].SDM are used in a large selection of topics: forest landscape [32], wetland distribution [33], coastal benthic biodiversity [34], medicine [35], aquatic invasive species [36, 37].
In earlier work, Diarra et al. [23] modelled the spatial distribution of five Culicoides species of veterinary interest using two statistical approaches: a generalized linear model (GLM) with Poisson distribution, and a random forest (RF) model. The choice of the species (C. imicola, C. oxystoma, C. enderleini, C. bolitinos and C. miombo) was justified by their vectorial competence for the BTV and AHS viruses [38–41].
In this study, we combined an Ecological Niche Factor Analysis (ENFA) [42, 43] and species distribution modelling. We used an ENFA to select variables contributing to the ecological niche. The main advantage of ecological niches models, compared to other traditional regression modelling approaches, is that they require only presence data [44] and they effectively assess the likelihood of species presence, or the relative ecological suitability of a spatial unit, within the study area [45].
We then used Boosted Regression Trees and MaxEnt to predict species distribution, and compared their results. These two methods are widely used species distributions models for Culicoides distribution [46] and for vector-borne diseases such as Rift Valley Fever (FVR) [47–50], Trypanosomosis [51, 52], Chikungunya [53, 54], Japanese Encephalitis Virus (JEV) [55, 56], Malaria [57–61], Epizootic Haemorrhagic Disease (EHD) [62], Dengue [63–65] and Plague [66, 67].
Our work completes that of Diarra et al. [23] investigating the potential effect of bioclimatic variables and livestock densities to predict spatial distribution for four Culicoides species that are potential vectors of BTV (C. imicola, C. oxystoma, C. enderleini and C. miombo), and to identify the most suitable habitats in Senegal.
Results
In all 1,373,929 specimens of the genus Culicoides belonging to at least 32 different species [23] were collected from 96 of the 108 sites visited at the end of the 2012 rainy season (in September and October). C. oxystoma, C. enderleini, C. imicola and C. miombo were the four most abundant species out of the species of veterinary interest [23]. For the 96 sites visited, C. oxystoma was present in 91 (94.79%), C. enderleini in 92 (95.83%), C. imicola in 92 (95.83%) and C. miombo in 77 (80.21%).
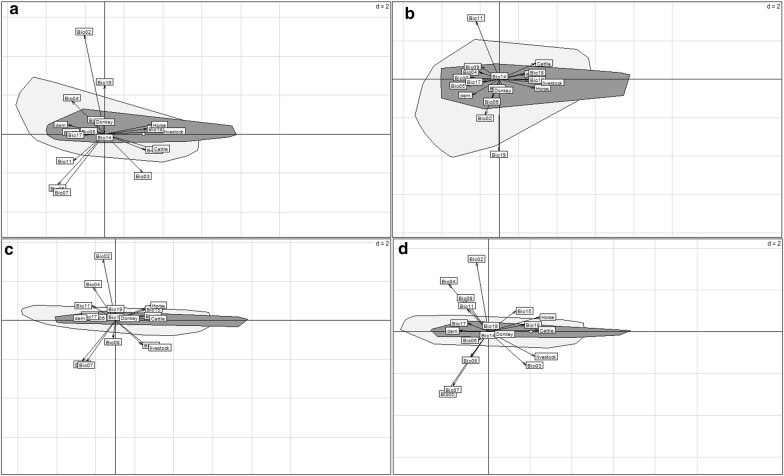
The ENFA (Fig. 1) showed that the presence of BTV vectors was often positively correlated to some precipitation variables, such as the precipitation of the warmest quarter (Bio18) and precipitation seasonality (Bio15), and to most of the livestock (horses, cattle, donkeys, goats and sheep), either cumulated or taken separately (Fig. 1a–d). On the other hand, the altitude (dem) and most of the temperature indices were negatively correlated to species occurrence, notably the maximum temperature of the warmest month (Bio05), mean temperature of the wettest quarter (Bio08) and the annual temperature range (Bio07) (Fig. 1).
Fig. 1.
Ecological niche factor analysis (ENFA) of Culicoides distribution in Senegal. C. imicola (a), C. oxystoma (b), C. enderleini (c) and C. miombo (d). Variables leading to ecological niche are represented into the light grey polygon and the dark grey polygon shows environmental conditions where Culicoides were observed (representation of the realized niche), and the small white circle corresponds to the barycentre of its distribution
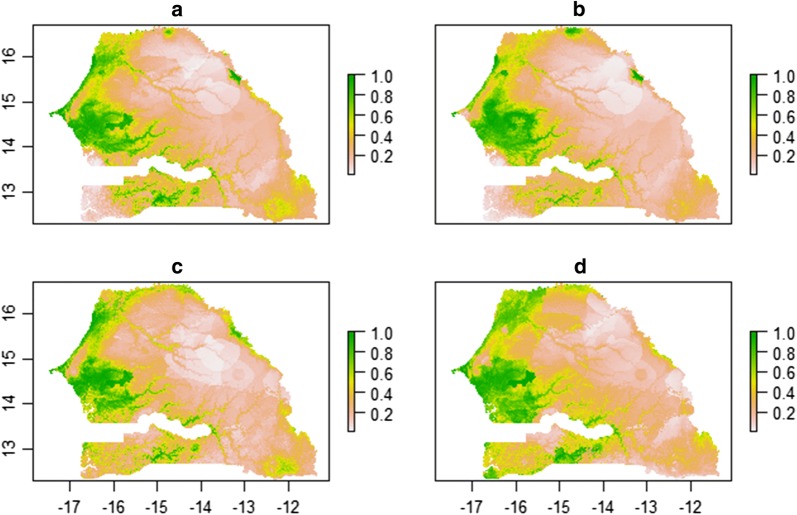
For each species we informed the MaxEnt (Fig. 2) and BRT (Fig. 3) models with variables previously found in the ENFA, to predict their geographical distribution. The resulting maps, showed the predicted geographical distributions of these species based on the habitat suitability for each of the four species. The green areas shown are those of greater relative probability of occurrence, while lighter coloured areas are those where the relative probability of occurrence was slight or null. For the MaxEnt model, a high probability of species presence was predicted in the Niayes area and the Groundnut Basin. The Niayes area and northern zone were predicted to be favourable for C. imicola (Fig. 2a), C. oxystoma (Fig. 2b), C. enderleini (Fig. 2c). For C. imicola, C. oxystoma, C. enderleini and C. miombo (Fig. 2d) the predicted presence probabilities were high from the northern Gambia to the eastern Senegal. In the southern Senegal, all species were predicted to be present. The probability of species occurrence was low in the Ferlo area and southeastern area. For each species, the corresponding niche model has an Area Under the Curve (AUC) greater than 0.77 (Table 1): 0.821 for C. imicola, 0.773 for C. oxystoma, 0.823 for C. enderleini and 0.785 for C. miombo.
Fig. 2.
MaxEnt predicted suitable areas. C. imicola (a), C. oxystoma (b), C. enderleini (c) and C. miombo (d). Green areas indicate areas that are likely to have suitable habitats for this vector species, while lighter areas indicate areas that are less suitable for the vector
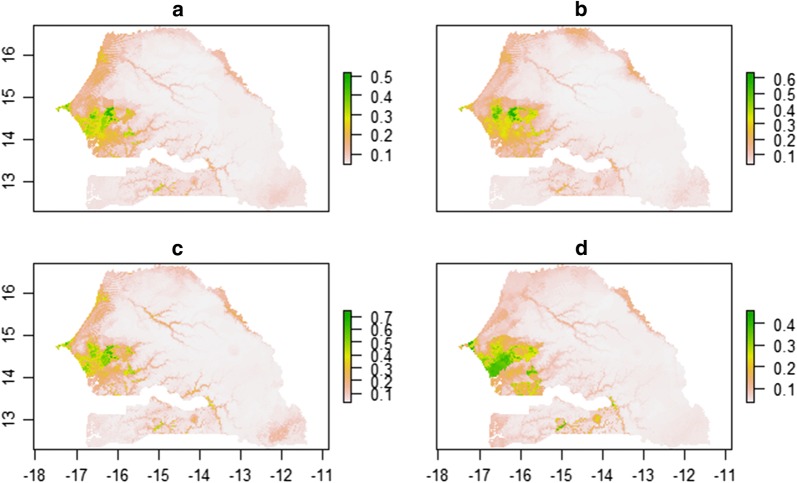
Fig. 3.
BRT predicted suitable areas. C. imicola (a), C. oxystoma (b), C. enderleini (c) and C. miombo (d). Green areas indicate areas that are likely to have suitable habitat for this vector species, while lighter areas indicate areas that are less suitable for the vector
Table 1.
Accuracy of the Niche Models: Area Under the Curve (AUC) for the MaxEnt and BRT models
| Species | MaxEnt AUC | BRT AUC |
|---|---|---|
| C. imicola | 0.821 | 0.813 |
| C. oxystoma | 0.773 | 0.817 |
| C. enderleini | 0.823 | 0.793 |
| C. miombo | 0.785 | 0.779 |
Comparatively to the MaxEnt model, the BRT model showed a similar predictive area for the ecological niche (Fig. 3). However, the probabilities of presence predicted by the BRT model were lower than those predicted by the MaxEnt model. The AUC values of the four species were greater than 0.77 (Table 1): 0.813 for C. imicola, 0.817 for C. oxystoma, 0.793 for C. enderleini and 0.779 for C. miombo.
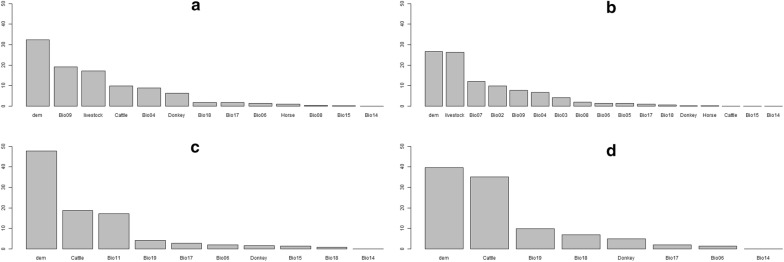
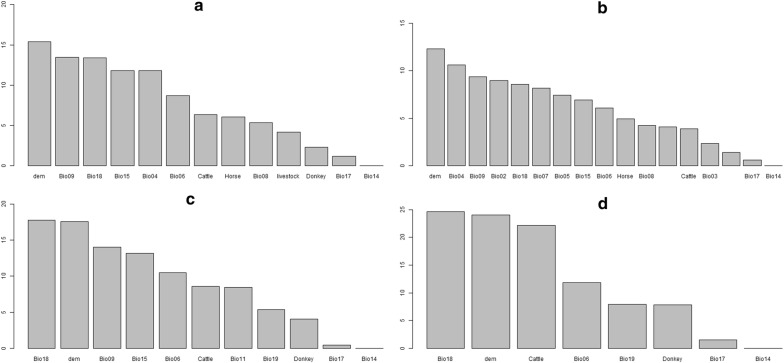
Figures 4 and 5 show the contributions of each of the environmental and livestock layers to the habitat suitability of the MaxEnt and BRT models, along with their influence.
Fig. 4.
Contribution (%) of each variable to the building of the Maxent models. C. imicola (a), C. oxystoma (b), C. enderleini (c) and C. miombo (d)
Fig. 5.
Contribution (%) of each variable to the building of the BRT models. C. imicola (a), C. oxystoma (b), C. enderleini (c) and C. miombo (d)
For the MaxEnt model, altitude was the most important variable driving Culicoides species distribution, all species included (Fig. 4). The other most important variables were the mean temperature of the driest quarter, the cumulative livestock density and temperature seasonality for C. imicola (Fig. 4a), the cumulated livestock density, annual temperature range, mean diurnal range and mean temperature of the driest quarter for C. oxystoma (Fig. 4b), the cattle density, mean temperature of the coldest quarter, precipitation of the coldest quarter and precipitation of the driest quarter for C. enderleini (Fig. 4c) and cattle density, mean temperature of the coldest quarter, precipitation of the warmest quarter, precipitation of the wettest quarter and donkey density for C. miombo (Fig. 4d).
Comparatively, for the BRT model, altitude was the most important variable driving Culicoides species distribution for two species, C. imicola and C. oxystoma and the precipitation of the warmest quarter for C. enderleini and C. miombo (Fig. 5). The other most important variables were the mean temperature of the driest quarter, precipitation of the warmest quarter, precipitation seasonality and temperature seasonality for C. imicola (Fig. 5a), temperature seasonality, the mean temperature of the driest quarter, mean diurnal temperature range and precipitation of the warmest quarter for C. oxystoma (Fig. 5b), the altitude, mean temperature of the driest quarter, precipitation seasonality and minimum temperature of the coldest month for C. enderleini (Fig. 5c), and the altitude, cattle density, minimum temperature of the coldest month and precipitation of the coldest quarter for C. miombo (Fig. 5d).
Hence, considering these two models, the most common contributing variables for building them were the altitude derived from digital elevation model (dem), the maximum temperature of the warmest month, the precipitation of the warmest quarter, mean temperature of the wettest quarter, temperature seasonality, the precipitation of the wettest quarter and the livestock density.
Discussion
Predictive modelling of species geographical distributions based on the environmental conditions of known occurrence sites is a major technique in analytical biology, with applications in conservation and reserve planning, ecology, evolution, epidemiology, invasive-species management and other fields [18–21].
A nationwide entomological sampling campaign enabled the collection of 1,373,929 specimens of the genus Culicoides belonging to at least 32 different species, at 96 different sites in 12 out of 14 regions of Senegal. For security reasons in southern Senegal, the Ziguinchor and Sédhiou regions were not visited.
In this study, ecological niche models were developed for four potential BTV vectors (C. imicola, C. oxystoma, C. enderleini and C. miombo [23]) using entomological data, climate, altitude variables and livestock density, to assess the effect of bioclimatic, altitude and livestock density variables on habitats suitable for Culicoides. The ENFA showed that the presence of BTV vectors was positively correlated to precipitation variables and to most of the livestock densities for all species, whilst the altitude (height) https://en.wikipedia.org/wiki/Digital_elevation_model and most of the temperature indices were negatively correlated to species occurrence. The MaxEnt and BRT models predicted the distribution of Culicoides based on the factors selected by ENFA. The two types of models used the same set of variables, but the importance of each of them varied depending on the species. The models predicted the same suitable zones, but with different probabilities of species presence. Each model had an AUC greater than 0.77. Based on the AUC, the MaxEnt was better than the BRT model for C. imicola, C. enderleini and C. miombo.
Temperature and precipitation are well known to be climate parameters that influence density and presence of Culicoides [68–72]. In this paper, the maximum temperature of the warmest month, the precipitation of the warmest quarter, the mean temperature of the wettest quarter, temperature seasonality and the precipitation of the wettest quarter were among the most driving factors for Culicoides species. In Senegal, the warmest and wettest months are during the rainy season (July–November), which includes the wettest quarter (August–October) and the warmest one (July–September). In addition, previous studies showed that the peak abundance of most Culicoides species is observed at the end of the rainy season (September–October) [73].
Furthermore, although each species has its own ecological requirements, any larval habitat could be shared by several ecologically-close species [17, 70]. This might explain the spatial co-occurrence of Culicoides species as seen in Fig. 2. Despite the fact that the presence of watercourses was not included as a predictor in our analysis, our model predicted the presence of Culicoides around Senegalese watercourses and lakes as expected [70]. Variations in Culicoides density are directly related to rainfall, hygrometry and temperatures, which condition the productivity of larval habitats and the spatial dispersion of adults [71, 74–76].
Our observations were consistent with those made by Diarra et al. [23]. In fact, by using two different statistical approaches, Random Forest (RF) and Generalized Linear models (GLM), Diarra et al. [23] showed that rainfall and/or NDVI were the most important variables influencing abundance for the 3 species C. imicola, C. enderleini and C. miombo. According to Diarra et al. [23], the abundance of C. oxystoma was mostly determined by the average rainfall and daily average temperature, that of C. enderleini by average precipitation, the Normalized Difference Vegetation Index (NDVI, a proxy for vegetation productivity) and the average daily temperature, that of C. imicola was mostly driven by the average precipitation and maximum NDVI, and that of C. miombo by NDVI followed by the average precipitation and average night temperature. As vegetation productivity in the Sahel zone is largely determined by climatic conditions, especially rainfall, we can, like Diarra et al. [23], confirm that variations in temperature and precipitation are among the best predictors of Culicoides occurrence and abundance. In contrast to Diarra et al. [23], we used new statistical approaches in this study with bioclimatic variables (19) covering a 50-year period, an altitude variable, and six more recent animal density variables. This gave 26 combinations of the four major variables (precipitation, temperature, altitude and livestock).
On the other hand, the ENFA showed that the occurrence Culicoides vectors of BTV was negatively correlated with altitude, which was the most important driver according to the MaxEnt and BRT models.
It is known that low-lying areas are often characterized by the presence of watercourses, and dense aquatic vegetation with a particular microclimate, so they are very suitable areas for livestock breeding and the development of arbovirus insect vectors [6, 7, 23, 73, 77]. Studies undertaken in South Korea [78], and in Kagoshima, southern Japan [79] showed the presence and abundance of Culicoides spp. in areas mainly characterized by a humid subtropical climate. Culicoides presence was also negatively correlated to variables linked to temperature, particularly the maximum temperature of the warmest month and the annual temperature range. Areas with these environmental conditions are predominantly covered by a low vegetation mantle (small bushes and trees). Thus, the Niayes area, where this type of vegetation is predominant, could be a suitable habitat for Culicoides species. Moreover, Diarra et al. [73], Fall et al. [80] and Diarra et al. [23] showed that C. oxystoma is very frequent and abundant in the Niayes area.
Livestock density was found to be positively associated with the occurrence and abundance of BTV vectors. Other studies [7, 81, 82] pointed in the same direction, showing the very complex relationship between Culicoides and their favourite hosts, ruminants and horses. For all species of interest, the Niayes area and the Groundnut basin were found to be the most suitable habitats, predicted with a high relative probability of occurrence (p > 0.7). In fact, both these areas are low-lying altitude and characterized by dense vegetation and high livestock density.
Furthermore, other potential predictor variables could be included in our model: the application of Culicoides control strategies, socio-economic status, human population densities, the presence of Culicoides biological predators, etc. However, our model effectively described habitat suitability using only altitude, temperature, precipitation and livestock density variables.
Conclusions
We presented ecological niche models for the BTV vectors, C. imicola, C. oxystoma, C. miombo and C. enderleini, on a nationwide scale in Senegal. This modelling approach allowed us to determine the effect of bioclimatic variables and to generate occurrences of Culicoides as risk factors for disease outbreaks. The results from this analysis can be used to (i) improve the quality of BT intervention plans identifying the areas of highest priority for intervention (reducing personnel and equipment costs) and (ii) provide a useful tool for researchers and disease control teams for further studies. Our models represent one of the first essential, albeit laborious, steps towards these future applications.
Materials and methods
Study area
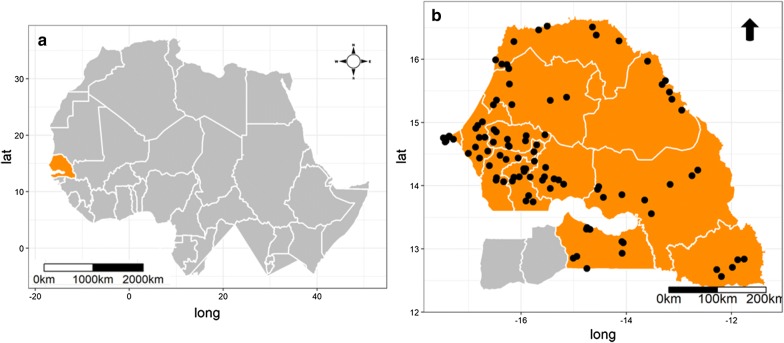
As a part of the nationwide surveillance programme in Senegal in 2012, 108 livestock premises were initially selected (as trapping sites) as follows: 3 departments per region, in 12 of the 14 Senegalese regions and 3 sites per department. The Ziguinchor and Sédhiou regions were excluded for safety reasons. In this study, we only considered data from 96 sites (Fig. 6) which were visited at the end of the 2012 rainy season (in September and October).
Fig. 6.
Map of Senegal, a West African country (a), with the location of study sites in 12 Senegalese regions (b). In yellow, the study area and in grey, the unsampled area
Data collection
Entomological data
Culicoides specimens were collected on two consecutive nights at each site using Onderstepoort black-light suction traps (Onderstepoort Veterinary Institute South Africa) positioned close to livestock’s enclosures. The geographical coordinates of each site were recorded with a Garmin© hand-held global positioning system receiver (accurate to within 10 m) and projected in UTM Zone 28N. Several identification keys were used depending on the species found and their subgenus or group [83–87]. For species that were difficult to identify, the specimens were dissected and slide-mounted in accordance with the Wirth and Marston technique for observation under a microscope [88, 89].
Climatic, environmental and livestock parameters
Several variables (26 in total) were used to implement the model. These were grouped in 4 categories (Table 2): 11 bio-climatic variables related to temperature (Bio01–Bio11); 8 bio-climatic variables related to precipitation (Bio12–Bio19); elevation data (1 variable) and animal density (6 variables).
Table 2.
Variables, description and code used in the ENFA and MaxEnt niche models
| Category of variables | Description | Abbreviation/code |
|---|---|---|
| Temperature | Mean annual temperature | Bio01 |
| Mean Diurnal range [mean of monthly (max temp − min temp)] | Bio02 | |
| Isothermality (Bio02/Bio07)*100 | Bio03 | |
| Temperature seasonality (standard deviation * 100) | Bio04 | |
| Maximum temperature of warmest month | Bio05 | |
| Minimum temperature of coldest month | Bio06 | |
| Annual temperature range (Bio05–Bio06) | Bio07 | |
| Mean temperature of wettest quarter | Bio08 | |
| Mean temperature of driest quarter | Bio09 | |
| Mean temperature of warmest quarter | Bio10 | |
| Mean temperature of coldest quarter | Bio11 | |
| Precipitations | Annual precipitation | Bio12 |
| Precipitation of wettest month | Bio13 | |
| Precipitation of driest month | Bio14 | |
| Precipitation seasonality (coefficient of variation) | Bio15 | |
| Precipitation of wettest quarter | Bio16 | |
| Precipitation of driest quarter | Bio17 | |
| Precipitation of warmest quarter | Bio18 | |
| Precipitation of coldest quarter | Bio19 | |
| Altitude | Digital elevation model | Dem |
| Animal density | Cumulative density of horses, cattle, donkeys, goats and sheep | Livestock |
| Cattle density | Cattle | |
| Goat density | Goat | |
| Sheep density | Sheep | |
| Horse density | Horse | |
| Donkey density | Donkey |
The bioclimatic data, with a spatial resolution of 30 arc-seconds (~ 1 km), were downloaded from the World Climate [90] website (http://www.worldclim.org/current) and averaged over a 50-year time period between 1950 and 2000 at the same spatial resolution. Elevation data (digital elevation model) were extrapolated from the Moderate Resolution Imaging Spectroradiometer (MODIS) with a spatial resolution of 30 arc-seconds (~ 1 km). Lastly, livestock data (number of head of cattle, small ruminant, horses and donkeys) were extracted from a survey undertaken at department level by the Direction des Services Vétérinaires (DSV), the Senegalese national institute and relevant body for animal health (DSV, 2013, unpublished work). Before being stacked together, the livestock data (6 variables) were rasterized by department with a spatial resolution of 250 m × 250 m. Livestock data were used due their importance for vector life [7, 81]. All layers were resampled at a spatial resolution of 250 m × 250 m using the nearest neighbour method and were stacked in one single spatial layer.
Modelling habitat suitability
We first carried out an exploratory analysis to identify the variables determining habitat suitability (i.e. the presence of the species). To that end, we used an Ecological Niche Factor Analysis (ENFA) [42, 43], a multivariate approach, similar to PCA, which requires only presence data for the species, in order to explore relationships between occurrence and environmental data. The first axis (marginality axis) of the ENFA is a measurement capturing the dimension in the ecological space conditions where the species is found are far from the global environmental conditions; a large marginality value would imply that the conditions where the species is found are “far” from the overall environmental conditions. In contrast, the second axis (specialization) is a measurement of the narrowness of the niche (ratio of the multidimensional variances of available to occupied spaces). During this procedure, highly correlated variables (with a coefficient of correlation higher than 0.95), and those that did not contribute to the ecological niche, were not used in the statistical analyses.
After selecting the variables, we used the MaxEnt approach [19, 91, 92] to model Culicoides presence. MaxEnt is a machine learning technique based, as the term indicates, on the principle of maximum entropy for a probability distribution, given constraints on its momenta:
where corresponds to species presence probability in the ith cell. The method uses presence locations and environmental covariates for all cells in the study area. Data, used to inform the model, define moment constraints on the distribution, while covariates define the mean, variance etc. where species occur. The result is an estimation of the presence probability in each cell.
Like MaxEnt, Boosted Regression Tree Modelling (BRT) [93] is another machine learning technique. The BRT approach developed by Friedman [94] uses two algorithms [30]: regression trees [95] and a boosting technique [96]. Over the past few years, this technique has emerged as one of the most powerful methods for predictive data mining. Some implementations of these powerful algorithms allow them to be used for regression as well as classification problems, with continuous and/or categorical predictors. Basically, the algorithm computes a sequence of simple trees, where each successive tree is built from the prediction residuals of the preceding tree.
Entomological data (for each species) were randomly divided into two samples, training and testing, using the K-fold cross-validation method. Accordingly, the original data were divided into k samples, then one of the k samples was selected as the validation set and the other k − 1 samples were the learning set. The performance score was calculated, and the operation was repeated by selecting another validation sample from the k − 1 samples that had yet used for model validation. The operation was repeated k times so that ultimately each sub-sample had been used exactly once as a validation set. The mean of the k mean squared errors was finally calculated to estimate prediction error. In this work we used k = 5.
Model performance was tested using the Area under the ROC curve (AUC), a plot of sensitivity against specificity measuring the ability of the model to discriminate between sites where a species was present (y = 1), as opposed to where it is absent (y = 0) [97–99]. AUC values range from 0 to 1; an AUC value higher than 0.8 indicates robust performance of the model. Statistical analysis and modelling were performed with R [100] using the following R-packages: adehabitatHS [101] for ENFA computation, dismo [102, 103] for MaxEnt and GBM [93] for BRT modelling.
Acknowledgements
The authors are grateful to all the people who provided assistance in operating traps on several nights around the trapping sites. This study was partially funded by EU grant FP7-261504 EDENext (http://www.edenext.eu) and data analysis was funded by the EU project ‘Understanding pathogen, livestock, environment interactions involving bluetongue’ (PALE‐BLU, H2020‐SFS‐2016‐2) through a PhD recruitment. The contents of this publication are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission. The authors are grateful to Fallou Niakh for Boosted Regression Tree statistical analyses. The authors are very thankful to Peter Biggins for English revisions. Finally, the authors are grateful to the anonymous reviewers for their helpful comments.
Abbreviations
- ENFA
ecological niche factor analysis
- AHS
African horse sickness
- AHSV
African horse sickness virus
- BT
bluetongue
- BTV
bluetongue virus
- AUC
area under the ROC curve
Authors’ contributions
MC, BB, AGF, MF and AA designed the study. BB, MC, AGF and MF collected the data. MF identified the species. BB, MC, AGF and AA performed statistical analyses. BB and MC wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Funding
The sampling phase was partially funded by the EU grant FP7-261504 EDENext and the Institut Sénégalais de Recherches Agricoles (ISRA) and data analysis was funded by the EU project ‘Understanding pathogen, livestock, environment interactions involving bluetongue’ (PALE‐BLU, H2020‐SFS‐2016‐2). The funding body did not play any roles in the design of the study and collection, analysis, and interpretation of data and in writing.
Availability of data and materials
All relevant data are presented or refer to their publicly available sources in the main text of the paper. Entomological data supporting the conclusions of this article are already published in Diarra et al. [23]. The bioclimatic data (Bio01–Bio19) used are available at the World Climate website: http://www.worldclim.org/current and the digital elevation model layer via the PALE-Blu data archive website in https://www.edenextdata.com/?q=content/modis-1km-digital-elevation-model-and-landwater-mask-v5
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mamadou Ciss, Email: ciss.mamadou@gmail.com.
Biram Biteye, Email: biteye88@yahoo.fr.
Assane Gueye Fall, Email: assane.fall@isra.sn.
Moussa Fall, Email: moussafall08@yahoo.fr.
Marie Cicille Ba Gahn, Email: mariececille.gahn@gmail.com.
Louise Leroux, Email: louise.leroux@cirad.fr.
Andrea Apolloni, Email: andrea.apolloni@cirad.fr.
References
- 1.Akakpo A, Wombou Toukam C, Mankor A, Ly C: Economic impact of african horse sickness outbreak in senegal in 2007. Inter-african bureau for animal resources po Box, NAIROBI, KENYA 2011, 59(1):1–16.
- 2.Diouf N, Etter E, Lo M, Lo M, Akakpo A. Outbreaks of African horse sickness in Senegal, and methods of control of the 2007 epidemic. Vet Rec. 2013;172(6):152. doi: 10.1136/vr.101083. [DOI] [PubMed] [Google Scholar]
- 3.Lefèvre P-C, Calvez D. La fièvre catarrhale du mouton (bluetongue) en Afrique intertropicale: influence des facteurs écologiques sur la prévalence de l’infection. Revue d’élevage et de médecine vétérinaire des pays tropicaux. 1986;39(3–4):263–268. [PubMed] [Google Scholar]
- 4.Lefèvre P-C, Taylor WP. Situation épidémiologique de la fièvre catarrhale du mouton (blue tongue) au Sénégal. Revue d’Elevage et de Médecine vétérinaire des Pays tropicaux. 1983;36(3):241–245. [PubMed] [Google Scholar]
- 5.Bakhoum MT: Ecologie et taxonomie intégrative des moucherons piqueurs du genre# Culicoides# Latreille (Diptera: Ceratopogonidae) en région Afrotropicale. PhD, N° 2017AGPT0012, 166 p, AgroParisTech, Paris, France. AgroParisTech; 2017.
- 6.Fall M, Fall AG, Seck MT, Bouyer J, Diarra M, Balenghien T, Garros C, Bakhoum MT, Faye O, Baldet T. Circadian activity of Culicoides oxystoma (Diptera: ceratopogonidae), potential vector of bluetongue and African horse sickness viruses in the Niayes area, Senegal. Parasitol Res. 2015;114(8):3151–3158. doi: 10.1007/s00436-015-4534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fall M, Fall AG, Seck MT, Bouyer J, Diarra M, Lancelot R, Gimonneau G, Garros C, Bakhoum MT, Faye O. Host preferences and circadian rhythm of Culicoides (Diptera: Ceratopogonidae), vectors of African horse sickness and bluetongue viruses in Senegal. Acta Trop. 2015;149:239–245. doi: 10.1016/j.actatropica.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Birley M, Boorman J. Estimating the survival and biting rates of haematophagous insects, with particular reference to the Culicoides obsoletus group (Diptera, Ceratopogonidae) in southern England. J Anim Ecol. 1982;51:135–148. doi: 10.2307/4315. [DOI] [Google Scholar]
- 9.Holmes P, Birley M. An improved method for survival rate analysis from time series of haematophagous dipteran populations. J Anim Ecol. 1987;56:427–440. doi: 10.2307/5058. [DOI] [Google Scholar]
- 10.Zimmer J-Y, Haubruge E, Francis F. Review: larval ecology of Culicoides biting midges (Diptera: Ceratopogonidae) Biotechnol Agron Soc Environ. 2014;18(2):301–312. [Google Scholar]
- 11.Purse B, Carpenter S, Venter G, Bellis G, Mullens B. Bionomics of temperate and tropical Culicoides midges: knowledge gaps and consequences for transmission of Culicoides-borne viruses. Annu Rev Entomol. 2015;60:373–392. doi: 10.1146/annurev-ento-010814-020614. [DOI] [PubMed] [Google Scholar]
- 12.Lubega R, Khamala C. Larval habitats of common culicoides Latreille (Diptera, Ceratopogonidae) in Kenya. Bull Entomol Res. 1976;66(3):421–425. doi: 10.1017/S0007485300006829. [DOI] [Google Scholar]
- 13.González M, López S, Mullens BA, Baldet T, Goldarazena A. A survey of Culicoides developmental sites on a farm in northern Spain, with a brief review of immature habitats of European species. Vet Parasitol. 2013;191(1–2):81–93. doi: 10.1016/j.vetpar.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 14.Dipeolu O, Ogunrinade A. Species of Culicoides breeding on rocks and riverbanks in Nigeria. Ecol Entomol. 1976;1(4):267–274. doi: 10.1111/j.1365-2311.1976.tb01231.x. [DOI] [Google Scholar]
- 15.Blackwell A, Young M, Mordue W. The microhabitat of Culicoides impunctatus (Diptera: Ceratopogonidae) larvae in Scotland. Bull Entomol Res. 1994;84(3):295–301. doi: 10.1017/S0007485300032399. [DOI] [Google Scholar]
- 16.Jenkins A, Young M. Breeding sites of Culicoides midges in KwaZulu-Natal. S Afr J Anim Sci. 2012;40(5):510–513. [Google Scholar]
- 17.Bakhoum MT, Fall AG, Fall M, Bassene CK, Baldet T, Seck MT, Bouyer J, Garros C, Gimonneau G. Insight on the larval habitat of Afrotropical Culicoides Latreille (Diptera: Ceratopogonidae) in the Niayes area of Senegal, West Africa. Parasites Vectors. 2016;9(1):462. doi: 10.1186/s13071-016-1749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Model. 2006;190(3–4):231–259. doi: 10.1016/j.ecolmodel.2005.03.026. [DOI] [Google Scholar]
- 19.Phillips SJ, Dudík M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31(2):161–175. doi: 10.1111/j.0906-7590.2008.5203.x. [DOI] [Google Scholar]
- 20.Chow TE, Gaines KF, Hodgson ME, Wilson MD. Habitat and exposure modelling for ecological risk assessment: a case study for the raccoon on the Savannah River Site. Ecol Model. 2005;189(1–2):151–167. doi: 10.1016/j.ecolmodel.2005.04.001. [DOI] [Google Scholar]
- 21.Corsi F, Duprè E, Boitani L. A large-scale model of wolf distribution in Italy for conservation planning. Conserv Biol. 1999;13(1):150–159. doi: 10.1046/j.1523-1739.1999.97269.x. [DOI] [Google Scholar]
- 22.Bouyer J, Dicko AH, Cecchi G, Ravel S, Guerrini L, Solano P, Vreysen MJ, De Meeûs T, Lancelot R. Mapping landscape friction to locate isolated tsetse populations that are candidates for elimination. Proc Natl Acad Sci. 2015;112(47):14575–14580. doi: 10.1073/pnas.1516778112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diarra M, Fall M, Fall AG, Diop A, Lancelot R, Seck MT, Rakotoarivony I, Allène X, Bouyer J, Guis H. Spatial distribution modelling of Culicoides (Diptera: Ceratopogonidae) biting midges, potential vectors of African horse sickness and bluetongue viruses in Senegal. Parasite Vectors. 2018;11(1):341. doi: 10.1186/s13071-018-2920-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter G, Gillison AN, Winter J. DOMAIN: a flexible modelling procedure for mapping potential distributions of plants and animals. Biodivers Conserv. 1993;2(6):667–680. doi: 10.1007/BF00051966. [DOI] [Google Scholar]
- 25.Mahalanobis PC. On the generalized distance in statistics. In: Proceedings of the National Institute of Science of India; 1936.
- 26.Hastie TJ, Tibshirani RJ. Generalized additive models. London: Taylor and Francis; 1990. [Google Scholar]
- 27.Wood SN. Generalized additive models: an introduction with R. Boca Raton: Chapman and Hall/CRC; 2006. [Google Scholar]
- 28.Breiman L. Random forests. Machine learning. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 29.Vapnik V. Statistical learning theory. New York: Wiley; 1998. [Google Scholar]
- 30.Elith J, Leathwick JR, Hastie T. A working guide to boosted regression trees. J Anim Ecol. 2008;77(4):802–813. doi: 10.1111/j.1365-2656.2008.01390.x. [DOI] [PubMed] [Google Scholar]
- 31.Phillips SJ, Dudík M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31:161–175. doi: 10.1111/j.0906-7590.2008.5203.x. [DOI] [Google Scholar]
- 32.Altamirano A, Miranda A, Meli P, Dehennin J, Muys B, Prado M, Catalán G, Smith-Ramírez C, Bustamante-Sánchez M, Lisón F. Spatial congruence among indicators of recovery completeness in a Mediterranean forest landscape: implications for planning large-scale restoration. Ecol Indicators. 2019;102:752–759. doi: 10.1016/j.ecolind.2019.03.046. [DOI] [Google Scholar]
- 33.Xue Z, Zou Y, Zhang Z, Lyu X, Jiang M, Wu H, Liu X, Tong S. Reconstruction and future prediction of the distribution of wetlands in China. Earth’s Future. 2018;6(11):1508–1517. doi: 10.1029/2017EF000807. [DOI] [Google Scholar]
- 34.Peterson A, Herkül K, Torn K. Modeling coastal benthic biodiversity using georeferenced environmental data: mapping present and predicting future changes. J Coast Res. 2018;85(sp1):376–380. doi: 10.2112/SI85-076.1. [DOI] [Google Scholar]
- 35.Deconinck E, Zhang MH, Petitet F, Dubus E, Ijjaali I, Coomans D, Vander Heyden Y. Boosted regression trees, multivariate adaptive regression splines and their two-step combinations with multiple linear regression or partial least squares to predict blood–brain barrier passage: a case study. Anal Chim Acta. 2008;609(1):13–23. doi: 10.1016/j.aca.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez-Rey M, Consuegra S, Börger L. Garcia de Leaniz C: Improving Species Distribution Modelling of freshwater invasive species for management applications. PLoS ONE. 2019;14(6):e0217896. doi: 10.1371/journal.pone.0217896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papeş M, Havel JE, Vander Zanden MJ. Using maximum entropy to predict the potential distribution of an invasive freshwater snail. Freshwat Biol. 2016;61(4):457–471. doi: 10.1111/fwb.12719. [DOI] [Google Scholar]
- 38.Mellor PS, Osborne R, Jennings DM. Isolation of bluetongue and related viruses from Culicoides spp. in the Sudan. J Hyg (Lond) 1984;93(03):621–628. doi: 10.1017/S0022172400065190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meiswinkel R, Paweska JT. Evidence for a new field Culicoides vector of African horse sickness in South Africa. Prev Vet Med. 2003;60:243–253. doi: 10.1016/S0167-5877(02)00231-3. [DOI] [PubMed] [Google Scholar]
- 40.Dadawala AI, Biswas SK, Rehman W, Chand K, De A, Mathapati BS, Kumar P, Chauhan HC, Chandel BS, Mondal B. Isolation of Bluetongue Virus Serotype 1 from Culicoides vector Captured in Livestock Farms and Sequence Analysis of the Viral Genome Segment-2. Transboundary Emerg Dis. 2012;59(4):361–368. doi: 10.1111/j.1865-1682.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- 41.Venter GJ, Mellor PS, Paweska JT. Oral susceptibility of South African stock-associated Culicoides species to bluetongue virus. Med Vet Entomol. 2006;20(03):329–334. doi: 10.1111/j.1365-2915.2006.00635.x. [DOI] [PubMed] [Google Scholar]
- 42.Hirzel AH, Hausser J, Chessel D, Perrin N. Ecological-niche factor analysis: how to compute habitat-suitability maps without absence data? Ecology. 2002;83(7):2027–2036. doi: 10.1890/0012-9658(2002)083[2027:ENFAHT]2.0.CO;2. [DOI] [Google Scholar]
- 43.Basille M, Calenge C, Marboutin E, Andersen R, Gaillard J-M. Assessing habitat selection using multivariate statistics: some refinements of the ecological-niche factor analysis. Ecol Model. 2008;211(1–2):233–240. doi: 10.1016/j.ecolmodel.2007.09.006. [DOI] [Google Scholar]
- 44.Neerinckx SB, Peterson AT, Gulinck H, Deckers J, Leirs H. Geographic distribution and ecological niche of plague in sub-Saharan Africa. Int J Health Geogr. 2008;7(1):54. doi: 10.1186/1476-072X-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elith J, Graham CH, Anderson RP, Dudík M, Ferrier S, Guisan A, Hijmans RJ, Huettmann F, Leathwick JR, Lehmann A. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29(2):129–151. doi: 10.1111/j.2006.0906-7590.04596.x. [DOI] [Google Scholar]
- 46.Sloyer KE, Burkett-Cadena ND, Yang A, Corn JL, Vigil SL, McGregor BL, Wisely SM, Blackburn JK. Ecological niche modeling the potential geographic distribution of four Culicoides species of veterinary significance in Florida, USA. PLoS ONE. 2019;14(2):e0206648. doi: 10.1371/journal.pone.0206648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sallam MF, Al Ahmed AM, Abdel-Dayem MS, Abdullah MA. Ecological niche modeling and land cover risk areas for rift valley fever vector, culex tritaeniorhynchus giles in Jazan, Saudi Arabia. PLoS ONE. 2013;8(6):e65786. doi: 10.1371/journal.pone.0065786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mweya CN, Kimera SI, Kija JB, Mboera LE. Predicting distribution of Aedes aegypti and Culex pipiens complex, potential vectors of Rift Valley fever virus in relation to disease epidemics in East Africa. Infect Ecol Epidemiol. 2013;3(1):21748. doi: 10.3402/iee.v3i0.21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sindato C, Stevens KB, Karimuribo ED, Mboera LE, Paweska JT, Pfeiffer DU. Spatial heterogeneity of habitat suitability for Rift Valley fever occurrence in Tanzania: an ecological niche modelling approach. PLoS Negl Trop Dis. 2016;10(9):e0005002. doi: 10.1371/journal.pntd.0005002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Redding DW, Tiedt S, Lo Iacono G, Bett B, Jones KE. Spatial, seasonal and climatic predictive models of Rift Valley fever disease across Africa. Philos Trans R Soc B Biol Sci. 2017;372(1725):20160165. doi: 10.1098/rstb.2016.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dicko AH, Lancelot R, Seck MT, Guerrini L, Sall B, Lo M, Vreysen MJ, Lefrançois T, Fonta WM, Peck SL. Using species distribution models to optimize vector control in the framework of the tsetse eradication campaign in Senegal. Proc Natl Acad Sci. 2014;111(28):10149–10154. doi: 10.1073/pnas.1407773111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chikowore G, Dicko AH, Chinwada P, Zimba M, Shereni W, Roger F, Bouyer J, Guerrini L. A pilot study to delimit tsetse target populations in Zimbabwe. PLoS Negl Trop Dis. 2017;11(5):e0005566. doi: 10.1371/journal.pntd.0005566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richman R, Diallo D, Diallo M, Sall AA, Faye O, Diagne CT, Dia I, Weaver SC, Hanley KA, Buenemann M. Ecological niche modeling of Aedes mosquito vectors of chikungunya virus in southeastern Senegal. Parasite Vectors. 2018;11(1):255. doi: 10.1186/s13071-018-2832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nsoesie EO, Kraemer MU, Golding N, Pigott DM, Brady OJ, Moyes CL, Johansson MA, Gething PW, Velayudhan R, Khan K. Global distribution and environmental suitability for chikungunya virus, 1952 to 2015. Euro surveill Bull Eur sur les maladies transmissibles Eur commun Dis Bull. 2016 doi: 10.2807/1560-7917.ES.2016.21.20.30234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller RH, Masuoka P, Klein TA, Kim H-C, Somer T, Grieco J. Ecological niche modeling to estimate the distribution of Japanese encephalitis virus in Asia. PLoS Negl Trop Dis. 2012;6(6):e1678. doi: 10.1371/journal.pntd.0001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Longbottom J, Browne AJ, Pigott DM, Sinka ME, Golding N, Hay SI, Moyes CL, Shearer FM. Mapping the spatial distribution of the Japanese encephalitis vector, Culex tritaeniorhynchus Giles, 1901 (Diptera: Culicidae) within areas of Japanese encephalitis risk. Parasites Vectors. 2017;10(1):148. doi: 10.1186/s13071-017-2086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moffett A, Shackelford N, Sarkar S. Malaria in Africa: vector species’ niche models and relative risk maps. PLoS ONE. 2007;2(9):e824. doi: 10.1371/journal.pone.0000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kulkarni MA, Desrochers RE, Kerr JT. High resolution niche models of malaria vectors in northern Tanzania: a new capacity to predict malaria risk? PLoS ONE. 2010;5(2):e9396. doi: 10.1371/journal.pone.0009396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akpan GE, Adepoju KA, Oladosu OR, Adelabu SA. Dominant malaria vector species in Nigeria: modelling potential distribution of Anopheles gambiae sensu lato and its siblings with MaxEnt. PLoS ONE. 2018;13(10):e0204233. doi: 10.1371/journal.pone.0204233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altamiranda-Saavedra M, Arboleda S, Parra JL, Peterson AT, Correa MM. Potential distribution of mosquito vector species in a primary malaria endemic region of Colombia. PLoS ONE. 2017;12(6):e0179093. doi: 10.1371/journal.pone.0179093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J, Patil AP, Temperley WH, Gething PW, Kabaria CW, et al. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic précis. Parasites Vectors. 2010;3(1):117. doi: 10.1186/1756-3305-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sloyer KE, Burkett-Cadena ND, Yang A, Corn JL, Vigil SL, McGregor BL, Wisely SM, Blackburn JK. Ecological niche modeling the potential geographic distribution of four Culicoides species of veterinary significance in Florida, USA. PLoS ONE. 2019;14(2):e0206648. doi: 10.1371/journal.pone.0206648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Althouse BM, Ng YY, Cummings DAT. Prediction of dengue incidence using search query surveillance. PLOS Negl Trop Dis. 2011;5(8):e1258. doi: 10.1371/journal.pntd.0001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashby J, Moreno-Madriñán M, Yiannoutsos C, Stanforth A. Niche modeling of dengue fever using remotely sensed environmental factors and boosted regression trees. Remote Sens. 2017;9(4):328. doi: 10.3390/rs9040328. [DOI] [Google Scholar]
- 65.Fatima SH, Atif S, Rasheed SB, Zaidi F, Hussain E. Species distribution modelling of aedes aegypti in two dengue-endemic regions of Pakistan. Trop Med Int Health. 2016;21(3):427–436. doi: 10.1111/tmi.12664. [DOI] [PubMed] [Google Scholar]
- 66.Holt AC, Salkeld DJ, Fritz CL, Tucker JR, Gong P. Spatial analysis of plague in California: niche modeling predictions of the current distribution and potential response to climate change. Int J Health Geogr. 2009;8(1):38. doi: 10.1186/1476-072X-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hieronimo P, Meliyo J, Gulinck H, Kimaro DN, Mulungu LS, Kihupi NI, Msanya BM, Leirs H, Deckers JA. Integrating land cover and terrain characteristics to explain plague risks in Western Usambara Mountains, Tanzania: a geospatial approach. Tanzan J Health Res. 2014;16(3):207–218. doi: 10.4314/thrb.v16i3.7. [DOI] [PubMed] [Google Scholar]
- 68.Mellor P, Boorman J, Baylis M. Culicoides biting midges: their role as arbovirus vectors. Annu Rev Entomol. 2000;45(1):307–340. doi: 10.1146/annurev.ento.45.1.307. [DOI] [PubMed] [Google Scholar]
- 69.Herniman K, Boorman J, Taylor W. Bluetongue virus in a Nigerian dairy cattle herd: 1. Serological studies and correlation of virus activity to vector population. Epidemiol Infect. 1983;90(2):177–193. doi: 10.1017/s0022172400028849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zimmer J-Y, Haubruge E, Francis F, Bortels J, Simonon G, Losson B, Mignon B, Paternostre J, De Deken R, De Deken G. Breeding sites of bluetongue vectors in northern Europe. Vet Rec. 2008;162(4):131. doi: 10.1136/vr.162.4.131. [DOI] [PubMed] [Google Scholar]
- 71.Braverman Y, Chechik F. Air streams and the introduction of animal diseases borne on Culicoides (Diptera, Ceratopogonidae) into Israel. Rev Sci Tech Off Int Epiz. 1996;15(3):1037–1052. doi: 10.20506/rst.15.3.968. [DOI] [PubMed] [Google Scholar]
- 72.Murray M. The seasonal abundance of female biting-midges, Culicoides-brevitarsis Kieffer (Diptera, Ceratopogonidae), in coastal south-eastern Australia. Aust J Zool. 1991;39(3):333–342. doi: 10.1071/ZO9910333. [DOI] [Google Scholar]
- 73.Diarra M, Fall M, Fall AG, Diop A, Seck MT, Garros C, Balenghien T, Allène X, Rakotoarivony I, Lancelot R. Seasonal dynamics of Culicoides (Diptera: Ceratopogonidae) biting midges, potential vectors of African horse sickness and bluetongue viruses in the Niayes area of Senegal. Parasite Vectors. 2014;7(1):147. doi: 10.1186/1756-3305-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gibbs EPJ, Greiner EC. The epidemiology of bluetongue. Comp Immunol Microbiol Infect Dis. 1994;17(3–4):207–220. doi: 10.1016/0147-9571(94)90044-2. [DOI] [PubMed] [Google Scholar]
- 75.Kirkeby C, Bødker R, Stockmarr A, Lind P, Heegaard PM. Quantifying dispersal of European Culicoides (Diptera: Ceratopogonidae) vectors between farms using a novel mark-release-recapture technique. PLoS ONE. 2013;8(4):e61269. doi: 10.1371/journal.pone.0061269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sellers R, Gibbs E, Herniman K, Pedgley D, Tucker M. Possible origin of the bluetongue epidemic in Cyprus, August 1977. Epidemiol Infect. 1979;83(3):547–555. doi: 10.1017/s0022172400026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diarra M, Fall M, Lancelot R, Diop A, Fall AG, Dicko A, Seck MT, Garros C, Allène X, Rakotoarivony I. Modelling the abundances of two major Culicoides (Diptera: Ceratopogonidae) species in the Niayes area of Senegal. PLoS ONE. 2015;10(6):e0131021. doi: 10.1371/journal.pone.0131021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oem J-K, Chung J-Y, Kwon M-S, Kim T-K, Lee T-U, Bae Y-C. Abundance of biting midge species (Diptera: Ceratopogonidae, Culicoides spp.) on cattle farms in Korea. J Vet Sci. 2013;14(1):91–94. doi: 10.4142/jvs.2013.14.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yanase T, Matsumoto Y, Matsumori Y, Aizawa M, Hirata M, Kato T, Shirafuji H, Yamakawa M, Tsuda T, Noda H. Molecular identification of field-collected Culicoides larvae in the southern part of Japan. J Med Entomol. 2013;50(5):1105–1110. doi: 10.1603/ME11235. [DOI] [PubMed] [Google Scholar]
- 80.Fall M, Diarra M, Fall AG, Balenghien T, Seck MT, Bouyer J, Garros C, Gimonneau G, Allène X, Mall I. Culicoides (Diptera: Ceratopogonidae) midges, the vectors of African horse sickness virus–a host/vector contact study in the Niayes area of Senegal. Parasite Vectors. 2015;8(1):39. doi: 10.1186/s13071-014-0624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bakhoum MT, Fall M, Seck M, Gardes L, Fall A, Diop M, Mall I, Balenghien T, Baldet T, Gimonneau G. Foraging range of arthropods with veterinary interest: new insights for Afrotropical Culicoides biting midges (Diptera: Ceratopogonidae) using the ring method. Acta Trop. 2016;157:59–67. doi: 10.1016/j.actatropica.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 82.Garros C, Gardes L, Allene X, Rakotoarivony I, Viennet E, Rossi S, Balenghien T. Adaptation of a species-specific multiplex PCR assay for the identification of blood meal source in Culicoides (Ceratopogonidae: Diptera): applications on Palaearctic biting midge species, vectors of Orbiviruses. Infect Genet Evol. 2011;11:1103–1110. doi: 10.1016/j.meegid.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 83.Boorman J. Culicoides (Diptera: Ceratopogonidae) of the Arabian peninsula with notes on their medical and veterinary importance. Fauna of Saudi Arabia. 1989;10:160–224. [Google Scholar]
- 84.Cornet M, Brunhes J. Révision des espèces de Culicoides apparentées à C. schultzei (Enderlein, 1908) dans la région afrotropicale (Diptera, Ceratopogonidae) Bull Soc Entomol Fr. 1994;99(149):64. [Google Scholar]
- 85.Cornet M, Chateau R. The Culicoides of West Africa (second note). Species related to C. similis Carter, Ingram & Macfie, 1920 (Diptera, Ceratopogonidae) Cahiers ORSTOM, Serie Entomologie Medicale at Parasitologie. 1970;8(2):141–173. [Google Scholar]
- 86.Cornet M, Nevill E, Walker A. Note sur les Culicoides (Diptera, Ceratopogonidae) du groupe de C. milnei Austen, 1909, en Afrique orientale et australe. Cah ORSTOM Ser Ent Med Parasitol Off Rech Sci Tech Outre Mer 1974.
- 87.Meiswinkel R. Afrotropical Culicoides: C (Avaritia) miombo sp. nov., a widespread species closely allied to C. (A.) imicola Kieffer, 1913 (Diptera: Ceratopogonidae) Onderstepoort J Vet Res. 1913;1991:58. [PubMed] [Google Scholar]
- 88.Ronderos MM, Spinelli GR, Sarmiento P. Preparation and mounting of biting midges of the genus Culicoides Latreille (Diptera: Ceratopogonidae) to be observed with a scanning electron microscope. Trans Am Entomol Soc. 2000;126:125–132. [Google Scholar]
- 89.Wirth WW, Marston N. A method for mounting small insects on microscope slides in Canada balsam. Ann Entomol Soc Am. 1968;61(3):783–784. doi: 10.1093/aesa/61.3.783. [DOI] [Google Scholar]
- 90.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25(15):1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 91.Elith J, Phillips SJ, Hastie T, Dudík M, Chee YE, Yates CJ. A statistical explanation of MaxEnt for ecologists. Divers Distrib. 2011;17(1):43–57. doi: 10.1111/j.1472-4642.2010.00725.x. [DOI] [Google Scholar]
- 92.Merow C, Smith MJ, Silander JA., Jr A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography. 2013;36(10):1058–1069. doi: 10.1111/j.1600-0587.2013.07872.x. [DOI] [Google Scholar]
- 93.Ridgeway G. Generalized Boosted Models: A guide to the gbm package. Update. 2007;1(1):2007. [Google Scholar]
- 94.Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001;29:1189–1232. doi: 10.1214/aos/1013203451. [DOI] [Google Scholar]
- 95.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. Boca Raton: Chapman and Hall/CRC; 1984. [Google Scholar]
- 96.Friedman JH. Stochastic gradient boosting. Comput Stat Data Anal. 2002;38(4):367–378. doi: 10.1016/S0167-9473(01)00065-2. [DOI] [Google Scholar]
- 97.Pearson RG, Raxworthy CJ, Nakamura M, Townsend Peterson A. Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J Biogeogr. 2007;34(1):102–117. doi: 10.1111/j.1365-2699.2006.01594.x. [DOI] [Google Scholar]
- 98.Young N, Carter L, Evangelista P. A MaxEnt model v3. 3.3 e tutorial (ArcGIS v10). Fort Collins, Colorado; 2011. https://www.coloradoview.org/wp-content/coloradoviewData/trainingData/a-maxent-model-v8.pdf. Accessed 24 Oct 2019.
- 99.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 100.R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2017. http://www.R-project.org/. Accessed 24 Oct 2019.
- 101.Calenge C. The package “adehabitat” for the R software: a tool for the analysis of space and habitat use by animals. Ecol Model. 2006;197(3–4):516–519. doi: 10.1016/j.ecolmodel.2006.03.017. [DOI] [Google Scholar]
- 102.Hijmans RJ, Elith J. Species distribution modeling with R. R package version 08-11 2013.
- 103.Hijmans RJ, Phillips S, Leathwick J, Elith J. dismo: Species distribution modeling. R package version 0.9-3. 2013. http://CRAN.R-project.org/package=dismo. Accessed 24 Oct 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are presented or refer to their publicly available sources in the main text of the paper. Entomological data supporting the conclusions of this article are already published in Diarra et al. [23]. The bioclimatic data (Bio01–Bio19) used are available at the World Climate website: http://www.worldclim.org/current and the digital elevation model layer via the PALE-Blu data archive website in https://www.edenextdata.com/?q=content/modis-1km-digital-elevation-model-and-landwater-mask-v5