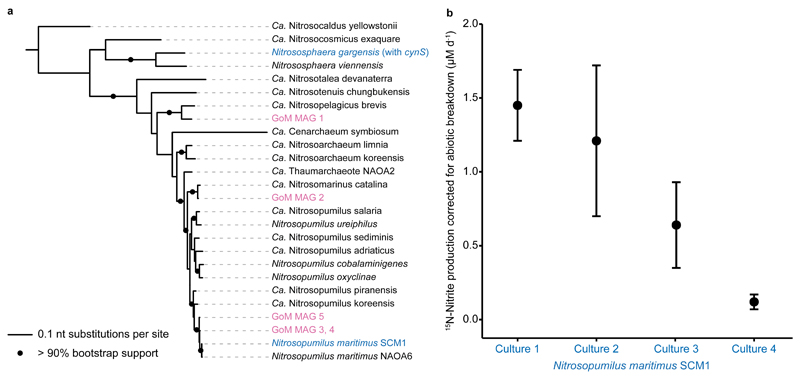
Fig. 4. amoA based phylogeny of Thaumarchaeota MAGs recovered in this study and cyanate utilization by the marine Thaumarchaeon Nitrosopumilus maritimus.
a) Phylogenetic placement of the amoA sequences from Gulf of Mexico metagenome assembled genomes (GoM MAGs, magenta) and Thaumarchaeota cultures that are able to utilize cyanate (blue). N. gargensis is the only Thaumarchaeon that encodes a known cyanase. Reference Thaumarchaeota amoA sequences are shown in black. Tree was constructed using IQ-TREE60 with automated model selection from near full-length amoA sequences and confidence was assessed with ultrafast bootstrapping (1,000 iterations). The scale bar represents nt substitutions per site, bootstrap support values >90% are depicted. b) 15N-nitrite production rate by the marine Thaumarchaeon N. maritimus SCM1 incubated with 15N-cyanate and a 14N-ammonium pool, corrected for abiotic breakdown of cyanate to ammonium in the culture medium. Error bars are the standard errors of the slope across all time points of one biological replicate. Rates were calculated based on linear regressions (one-sided t-test, t = 6.13, DF = 2, p = 0.012; t = 2.38, DF = 2, p = 0.070; t = 2.22, DF = 2, p = 0.078; t = 3.72, DF = 2, p = 0.033, for culture 1, 2, 3 and 4, respectively). When data were fitted with an exponential regression, p was <0.001 for all four cultures. Differences in rates between biological replicates correlate with the different starting biomass in each culture.