Fig. 2. BRN4 expression is selectively upregulated in CRPC-NE patient-derived xenograft (PDX) models, clinical samples and cell line models.
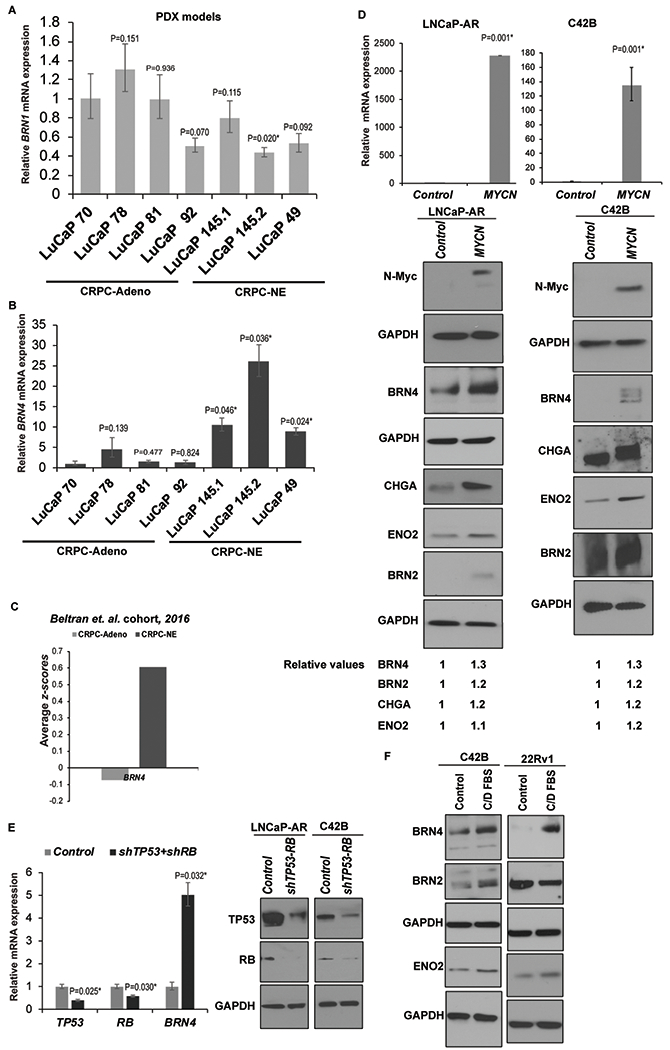
A. Relative BRN1 mRNA and;
B. BRN4 mRNA expression in PDX models with CRPC-Adenocarcinoma characteristics (LuCaP 70, 78, 81 and 92) vs those with CRPC-NE alterations (LuCaP 49, 145.1 and 145.2) as assessed by real-time PCR analyses. Data were normalized to GAPDH control and represented as mean ± SEM. P-values were calculated relative to LuCaP 70 PDX.
C. Average z-scores for BRN4 mRNA expression in CRPC-Adeno vs CRPC-NE tissues in Beltran et al (11) cohort.
D. LNCaP-AR and C42B cells were stably transfected with control/NMYC overexpression constructs followed by real time PCR analyses of NMYC mRNA (upper panel) and Western blot analyses of N-Myc, BRN4 and neuronal markers BRN2, CHGA and ENO2 (lower panels). Band intensities were quantified by Image J, relative values were calculated for indicated proteins after normalizing to corresponding GAPDH values and are represented below blots.
E. Left panel: LNCaP-AR cells were transfected with control shRNA/shRNA targeting TP53 and RB1 followed by real time PCR analyses of TP53, RB and BRN4 mRNA. Right panel: Western blot analyses confirming TP53 and RB knockdown following shRNA transfections. GAPDH was used as a loading control.
F. C42B and 22Rv1 cell lines were grown under androgen depleted conditions (RPMI medium with 10% C/D FBS) for 5 days followed by Western blot analyses of BRN4, BRN2 and indicated neuronal markers. GAPDH was used as a loading control.
