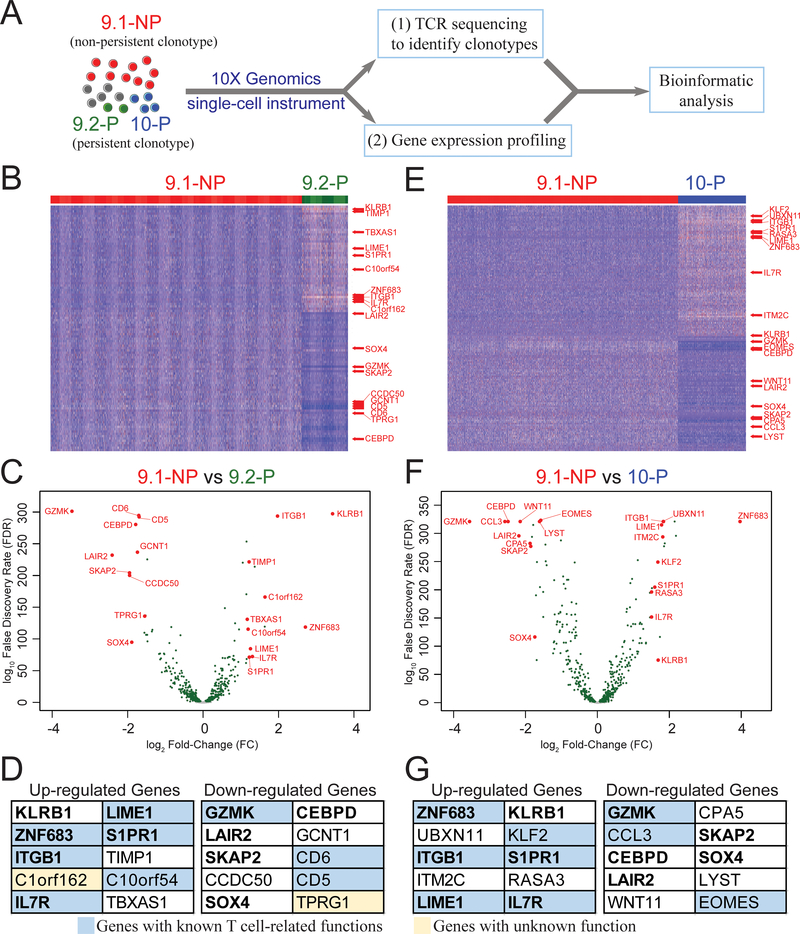
Figure 1. Persistent 9.2-P and 10-P single cells showed different gene expression profiles compared to the non-persistent 9.1-NP single cells.
A, TIL4095, contained three dominant clonotypes 9.1-NP, 9.2-P and 10-P (4). TIL4095 T cells were subjected to a single-cell instrument (10X Genomics) to perform (i) single-cell TCR sequencing and (ii) single-cell gene expression profiling. Each single cell contained a unique barcode, which could be used to link the gene expression profiles to one of the three dominant clonotypes (9.1-NP, 9.2-P or 10-P), based on their unique TCR-CDR3β sequences. This set of information was used in the subsequent analysis. Next, single cells from 9.1-NP non-persistent clonotype were compared with single cells from 9.2-P persistent clonotype to generate (B) a heatmap and (C) a volcano plot. (D) Top 10 upregulated genes and top 10 downregulated genes based on the fold changes (FC) are listed. Genes with known T cell-related functions are shown in blue boxes. Genes with unknown function are shown in yellow boxes. Single cells from 9.1-NP non-persistent clonotype were then compared with single cells from 10-P persistent clonotype to generate (E) a heatmap and (F) a volcano plot. (G) Top 10 upregulated genes and top 10 downregulated genes based on the FC are listed. Genes with known T cell-related functions are shown in blue boxes. For the heatmaps, each vertical line represents a single cell. Only genes with FC>1.5 and false discovery rate (FDR)<0.05 are shown. For the volcano plots, genes with FDR<0.05 are shown in green. The eleven genes shared between (D) and (G) are in bold text.