Abstract
Long non-coding RNA (lncRNA) prostate cancer associated transcript 1 (PCAT-1) has been identified as a potential biomarker for the diagnosis and prognosis of various cancers. We performed this systematic review and meta-analysis to evaluate the role of dysregulation as well as the biological and clinical significance of lnc-PCAT-1 for predicting the malignancy status in several cancers. Two independent reviewers conducted an extensive search in electronic databases of Medline, Embase, Scopus, Web of Science and PubMed until the December of 2017. Five articles investigating the clinical significance of lncRNA PCAT-1, including 996 patients, were analyzed. Our results revealed that the increased PCAT-1 expression was related to overall survival (OS) (HR = 1.9, 95% CI: 1.13-3.18, P=0.015). Also, pooled results of the diagnostic data analysis demonstrated that PCAT-1 has a sensitivity of 0.59 and specificity of 0.66 for cancer diagnosis. Moreover, pooled area under curve was 0.62 (95% CI: 0.58–0.69). This meta-analysis revealed that lncRNA PCAT-1 could be served as a potential diagnostic and prognostic biomarker in various solid tumors.
Keywords: Diagnosis, long non-coding RNA, PCAT-1, prognosis, survival
Introduction
Long non-coding RNAs (lncRNAs) are RNA transcripts characterized by a length about 200 nucleotides or more in length. These RNA molecules have been shown to regulate the gene expression at the transcriptional and post-transcriptional levels (Beermann et al., 2016; Parikshak et al., 2016; Adams et al., 2017; Qian et al., 2017).
Until recently, a number of studies have reported the aberrant expression of lncRNAs in various malignancies (Beermann et al., 2016; Parikshak et al., 2016; Adams et al., 2017; Qian et al., 2017). In the last decades, numerous studies have investigated the role of lncRNAs in cancer development and progression (Cao, 2014; Karlsson and Baccarelli., 2016). Accordingly, it has been shown that lncRNAs involve in a wide range of biological and cellular processes, including, cell growth, proliferation, differentiation, migration, invasion and apoptosis. Also, it has been demonstrated that lncRNAs are correlated with clinicopathological features in cancer patients and could be served as potential diagnostic and prognostic biomarkers (Wang et al., 2016; Chandra Gupta and Nandan Tripathi., 2017; Li et al., 2017).
Prostate cancer–associated lncRNA transcripts 1 (PCAT-1), encoded by the PCAT-1 gene mapped on chromosome 8q24.21, is a long intergenic non-coding RNA (Shi et al., 2015; Yan et al., 2015). This lncRNA firstly was annotated by RNA sequencing of prostate tissues and cells lines and transcriptome analysis (Prensner, Iyer et al. 2011). The functional studies confirmed that lncRNA PCAT-1 could promote cell proliferation through binding to polycomb repressive complex 2 (PCR2) as a transcriptional repressor (Saus et al., 2016).
Moreover, a number of clinical investigations promoted the diagnostic and prognostic value of PCAT-1 in various cancers. PCAT-1 has also been suggested as lncRNA that is related to overall survival (OS) in various carcinomas, including colorectal, gastric, breast, bladder, lung cancers, andmultiple myeloma, extrahepatic cholangiocarcinoma (ECC), hepatocellular carcinoma and esophageal squamous cell carcinoma (ESCC) (Ge et al., 2013; Shi et al., 2015; Yan et al., 2015; Qin et al., 2016; Cui et al., 2017). Altogether, PCAT-1 was supported that act as a prognostic marker and a therapeutic target for cancer therapy (Evans et al., 2014; Shi et al., 2015; Qiao et al., 2017; Shen et al., 2017; Zhang et al., 2017). Nevertheless, there are some limitations, including study design and insufficient sample size can be lead to inaccurate results. In addition, it is vital to investigate some important risk factors related to PCAT-1 in various malignancies.
Ever since, no meta-analysis study has been completed to explore the clinical value of PCAT-1 as a biomarker for various cancers. Thus, we carried out the present systematic review and meta-analysis study to evaluate the biological and clinical significance of PCAT-1 in several malignancies.
Biological significance
Experimental investigations revealed that PCAT-1 exerts a vital function in oncogenic signaling pathways related to cell proliferation. Recently, the experimental investigations revealed that lncRNA PCAT-1 can increase the proliferation, metastasis and invasion of cervical cancer cells (Ma et al., 2018).
The biological role of lncRNA PCAT-1 in the regulation of proto-oncogene c-Myc in prostate cancer has recently been reported. The tumorigenic effect of lnc-PCAT-1 in colorectal cancer (CRC) through inhibiting the c-Myc has been described (Qiao et al., 2017). Also, this lncRNA could promote CRC cell migration and invasiveness, as confirmed by the increased viability and induced apoptosis (Qiao et al., 2018). Altogether, it was suggested that PCAT-1 is an important lncRNA in the regulation of the c-Myc expression and corresponding proliferative signaling pathways (Evans et al., 2014).
In esophageal squamous cell carcinoma (ESCC), lnc-PCAT-1 has been demonstrated to promote cancer cell proliferation and growth, indicating it as a therapeutic target for ESCC (Zhen et al., 2018).
Moreover, the functional studies showed that lnc-PCAT-1 may increase cells proliferation, migration and invasion in osteosarcoma by interacting with EZH2 and repressing p21 gene. These findings suggest lnc-PCAT-1 as a oncogenic mediator in osteosarcoma carcinogenesis (Huang et al., 2018).
Clinical significance
The relationship between the dysregulation of PCAT-1 and the progression of carcinoma have been investigated in colorectal and gastric cancers, hepatocellular and esophageal squamous cell carcinoma (Ge et al., 2013; Shi et al., 2015; Yan et al., 2015; Qin et al., 2016; Cui et al., 2017). Moreover, the aberrant expression of this lncRNA was correlated with clinicopathological features in a number of malignancies, indicating it as a potential marker for early cancer detection (Shi et al., 2015; Yan et al., 2015; Qin et al., 2016; Cui et al., 2017). Dysregulation of lnc-PCAT-1 also designated as an independent prognostic factor for the overall survival (OS) rate of cancer patients (Shi et al., 2015; Yan et al., 2015; Qin et al., 2016; Cui et al., 2017).
Materials and Methods
Publication search
Two independent reviewers performed an extensive search in several electronic databases such as Springer, Medline, BioMed Central, ScienceDirect, Scopus, Embase, Web of Science, CNKI, CBM, Cochrane Central Register of Controlled Trials (CENTRAL) Cochrane Library, China National Knowledge Infrastructure and Chinese WanFang database for the published studies up to December 2017. Search strategy was based on the words presented in Table 1.
Table 1.
Queries Used in Medline, Embase and Scopus
| Database | Search terms |
|---|---|
| MEDLINE | ("cancer"[Mesh] OR "tumor"[tiab] OR "malignancy"[tiab] OR "neoplasm"[tiab] OR " adenocarcinoma"[tiab] AND ("long noncoding RNA"[MeSH Terms] OR "long non-coding RNA"[tiab] OR "Long non-coding RNA"[tiab] OR "Long noncoding RNAs"[tiab] OR " Long noncoding lncRNA"[tiab] OR " Long noncoding lncRNAs"[tiab] OR "long noncoding lncRNAs "[tiab] OR " long noncoding lncRNAs "[tiab] AND ("Prostate cancer-associated lncRNA transcripts 1"[MeSH Terms] OR "Prostate cancer-associated lncRNA transcripts 1"[tiab] OR "PCAT-1"[tiab] OR "PCAT1"[tiab] OR "plasma"[tiab] OR "serum"[tiab] OR "circulating long noncoding lncRNA "[tiab] OR " blood stream"[tiab]) |
| EMBASE | 1- exp" cancer"/ or ("tumor" OR "malignancy" OR "neoplasm "[tiab] OR "adenocarcinoma").ti,ab. |
| 2- exp oncoding lncRNA" OR "Long non-coding lncRNAs" OR " Long noncoding lncRNA" OR " Long noncoding lncRNAs" OR " long noncoding lncRNA "AND "Prostate cancer-associated lncRNA transcripts 1"/ or "PCAT1" OR "PCAT-1" OR "plasma" OR "serum" OR " long noncoding lncRNA " OR "blood stream").ti,ab. | |
| 3- 1 & 2 | |
| SCOPUS | ((TITLE-ABS-KEY (cancer) OR TITLE-ABS-KEY (adenocarcinoma) OR TITLE-ABS-KEY (malignany) OR TITLE-ABS-KEY (neoplasm) OR TITLE-ABS-KEY (plasma))) |
| AND ((TITLE-ABS-KEY (Prostate cancer-associated lncRNA transcripts 1) OR TITLE-ABS-KEY (PCAT-1) OR TITLE-ABS-KEY (PCAT1) AND((TITLE-ABS-KEY (serum) OR TITLE-ABS-KEY (blood stream) OR TITLE-ABS-KEY (plasma) OR TITLE-ABS-KEY (circulating) OR TITLE-ABS-KEY (lnc RNA) OR TITLE-ABS-KEY (cancer) OR TITLE-ABS-KEY (circulating lnc RNA) OR TITLE-ABS-KEY (plasma lncRNA) OR TITLE-ABS-KEY (circulation OR TITLE-ABS-KEY (circulating long noncoding RNA) OR TITLE-ABS-KEY (Long noncoding RNA) |
The applied search strategy was based on both medical subject heading terms and free-text words in order to enhance the sensitivity of the database search. The main keywords selected for the search were used as follows: “PCAT-1 and cancer”, “lnc-PCAT-1 and cancer”, “long non-coding RNA PCAT-1”, “lncRNA PCAT-1”, “PCAT-1”, “prostate cancer associated transcript 1”. Furthermore, all findings presenting as abstract in conferences were excluded from the study. The extraction and evaluation of relevant eligible studies were completed by two independent investigators.
In the meantime, all reference lists of related articles were also examined in order to conclude the eligible studies and also the recovered references were disregarded due to duplication. Moreover, the titles, abstracts, as well as full texts were carefully read to remove unrelated studies with no eligibility criteria.
Inclusion and exclusion criteria
Inclusion criteria: 1) A relationship of dichotomous PCAT-1 expression level with prognosis and diagnosis factors, clinical consequence in cancer patients; 2) A potential role of PCAT-1 specifically in the pathogenesis of any type of cancers; 3) A hazard ratios (HRs) and 95% its confidence intervals (CIs) for overall survival (Cox proportional hazard analysis, Kaplan–Meier method); 4) Published in English.
Exclusion criteria: 1) Studies of non dichotomous lncRNA PCAT-1 expression or absence of survival outcome; 2) Multiple duplicated articles; 3) Letters and conference abstracts.
Data analysis method
STATA version 14.0 (Stata Corporation, College Station, TX) was used for statistical analysis. Also, heterogeneity between the studies was performed by chi square and I2 tests and the level of significance for statistical tests was 0.05 based on Cochran-Q value (showing heterogeneity). We applied the fixed effect model when the data were homogeneous. When the cause of heterogeneity was not known, the random effect model was used. Lastly, the results of the studies were pooled and an overall effect size was given. It is worth noting that meta-analyses were performed when there are at least 3 studies related to reported data. Moreover, Begg’s test was carried out for recognizing publication bias.
Results
Biological and clinical relevance of lnc PCAT-1
Our focus was on the studies indicated lncRNA PCAT-1 as a potential effector in cellular process as well as its role as a clinical biomarker for several cancers. Table 2 shows the biological and clinical significance of lnc-PCAT-1 in a number of cancers.
Table 2.
Description of Potential Prognostic lncRNA Markers in Colorectal Cancer
| Type of cancer | Biological (Ref) | Clinical (Ref) |
|---|---|---|
| Hepatocellular carcinoma | Promoting cell invasion and metastasis through inhibiting miR-129-5p (Zhang et al., 2017). | Potential candidate for HCC diagnosis and treatment (Zhang et al., 2017). |
| Increasing cell proliferation and migration, decreasing apoptosis (Wen et al., 2016). | Candidate biomarker for HCC therapeutic strategies (Wen, Xu et al. 2016). | |
| Potential diagnostic tool or therapeutic target for HCC (Zhang et al., 2016). | ||
| Gastric cancer | Promoting cell proliferation, migration and invasion through regulating CDKN1A (Bi et al., 2017). | Potential diagnostic marker of gastric cancer (Bi et al., 2017). |
| Colorectal cancer | Promoting cell invasion, metastatic potential and drug resistance through c-Myc(Qiao et al., 2017). | Potential candidate biomarker for CRC (Qiao et al., 2017). |
| Increasing cell proliferation (Qiao et al., 2017). | ||
| Breast cancer | Correlating with clinical characteristics and as a biomarker for breast cancer (Sarrafzadeh et al., 2017). | |
| Prostate cancer | Promoting proliferation, migration and invasion, inhibiting apoptosis (Xu et al., 2017). | As a predictive biomarker (PCAT-1) was applied in PARP1 inhibitor therapy of patients (Prensner et al., 2014). |
| Increasing cell proliferation through modulating cMyc function (Evans et al., 2014). | As key aspects of disease biology and clinically important biomarkers (Prensner et al., 2011; Prensner et al., 2011). | |
| Acting as oncogenic lncRNA in prostatecancer proliferation through cMyc(Prensner, Chen et al., 2014). | ||
| Regulating BRCA2 levels (Prensner et al., 2011, Prensner et al., 2011) and homologous recombination (Prensner et al., 2014). | ||
| Lung cancer | Promoting cell proliferation, migration and invasion (Zhao et al., 2015). | Potential therapeutic target (Zhao et al., 2015). |
| Bladder cancer | Decreasing cell proliferation and increasing cell apoptosis (Liu et al., 2015). | Acting as a biomarker for predicting bladder cancer (Liu et al., 2015; Xu et al., 2017). |
| Cervical cancer | Increasing cell proliferation, metastasis and invasion (Ma et al., 2018). | |
| Osteosarcoma | Increasing cell proliferation, migration and invasion by interacting with EZH2 and repressing p21 gene (Huang et al., 2018). | |
| Eusophagus squamous cell carcinoma | Promoting cancer cell proliferation and growth (Zhen., 2018). | |
| Multiple myeloma | Correlating with the clinicopathologic features and acting as a potential diagnostic target (Shen et al., 2017). | |
| Extrahepatic Cholangiocarcinoma | Promoting cell proliferation and inducing cell apoptosis. Acting as a competing endogenous RNA (ceRNA) against miR-122. | |
| Regulating WNT1 expression via miR-122 (Zhang et al., 2017). |
CRC, colorectal cancer; EMT, Epithelial-mesenchymaltransition; lincRNA, long intergenic non-coding RNA, MM, Multiple myeloma; ECC, Extrahepatic cholangiocarcinoma; PARP-1, Poly [ADP-ribose] polymerase 1; NSCLC, Non-small-cell lung carcinoma.
The characteristics of the study
First, 1460 articles were extracted from electronic databases and after removing duplicate articles, 980 articles were screened according to a potential prognostic or diagnostic role of lncRNA PCAT-1 in cancers. Then, the full texts of 50 articles werereviewed and evaluated, 5 articles were finalized to meta-analysis because having aforementioned eligible inclusion criteria (Ge et al., 2013; Shi et al., 2015; Yan et al., 2015; Qin et al., 2016; Cui et al., 2017) (Figure 1). This Figure shows the stages of finding the appropriate articles for the current study.
Figure 1.
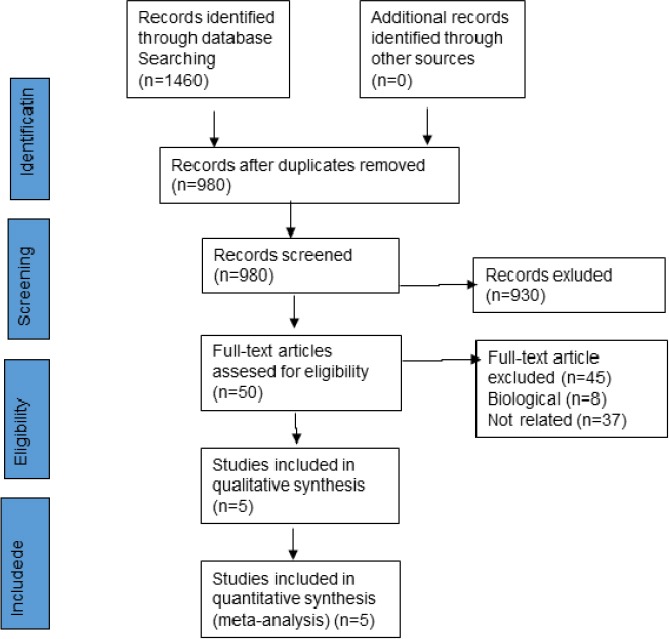
Flowchart of the Meta-Analysis
Relationship between lncRNA PCAT-1 expression and clinical outcome
A total of 996 patients from 5 studies were included for meta-analysis that all these studies were completed in Chinese population. Five different cancer types were including colorectal cancer (Ge et al., 2013), gastric cancer (Cui et al., 2017), hepatocellular carcinoma (Yan et al., 2015), Esophageal squamous cell carcinoma (Shi et al., 2015; Qin et al., 2016). The included cancer patients were classified into two groups (low/ high) based on the expression level of PCAT-1 for the median and ROC values.
Figure 2 shows the quality of the included studies has been assessed by QUADAS-2. As it is shown, all studied articles were on the upper middle quality. However, there is an obvious major bias in these included articles. It means that, there are clear weaknesses in the ‘‘index text,’’ which was 65% for these eligible studies.
Figure 2.
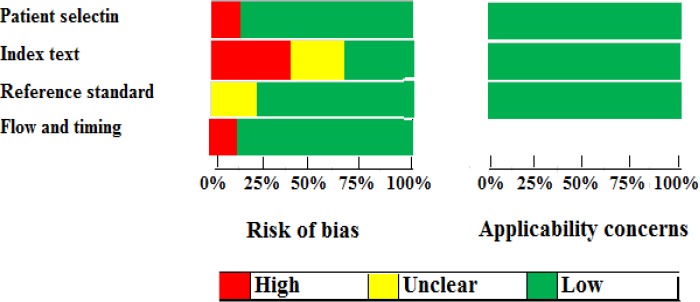
The Flow Diagram of Quality Assessment Using the QUADAS Check List Based on “Risk of Bias’’ and ‘‘Applicability Concerns”
Table 3 shows the main characteristics of patient cancers.
Table 3.
Main Characteristics of the Include Studies for Clinical Studies
| Study | Region | Tumor type | Sample size | Test method | Cut off | Outcome measure | HR estimation | Follow up Months |
|---|---|---|---|---|---|---|---|---|
| Ge 2013 | China | CRC | 189 | qRT-PCR | ROC | OS | Directly | ~90 |
| Yan 2015 | China | HCC | 117 | qRT-PCR | Median value | OS | Directly | ~60 |
| Shi 2015 | China | ESCC | 194 | qRT-PCR | Median value | OS | Directly | ~60 |
| Qin 2016 | China | ESCC | 321 | PCR, Sequencing | Median value | OS | Directly | ~60 |
| Cui 2017 | China | GC | 175 | qRT-PCR | Median value | OS | Directly | ~60 |
CRC, Colorectal cancer; HCC, Hepatocellular carcinoma; ESCC, Esophageal squamous cell carcinoma; GC, Gastric cancer; OS, overall survival; ROC, Receiver Operating Characteristic
Risk of Bias
In the present study there was no publication bias. According to Begg’s test (Z=-0.49; p=0.25) no publication bias was detected.
Meta-analysis results
The relationship between PCAT-1 and OS
Five articles, including 996 cancer patients were identified as selected studies in relationship with the expression level of PCAT-1 and overall survival (OS) (Figure 3). Because of heterogeneity among studies, the random effect model was applied to calculate the overall hazard (I2= 81.4%, P-value<0.001).
Figure 3.
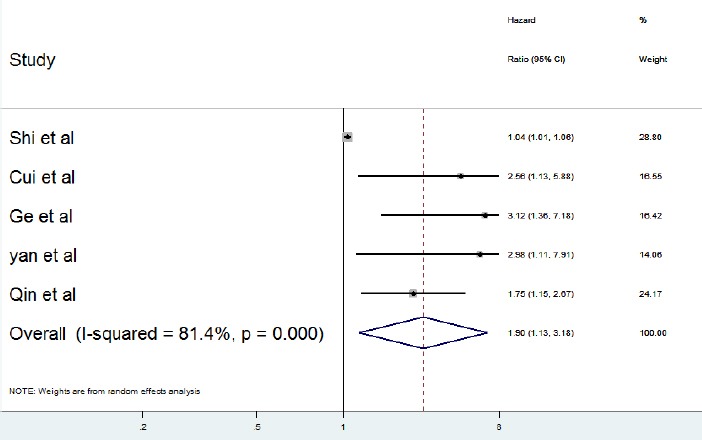
Forest Plot for the Relationships between PCAT-1 Expression and OS
The overall and pooled results revealed that the high expression level of PCAT1 was significantly related to shorter overall survival in cancer patients (HR =1.9, 95% CI: 1.13-3.18, P<0.001).
Diagnostic value of PCAT-1 expression
Five studies, including 996 cancer patients and 300 patients with low level of PCAT-1 expression and 551 patients with high level of PCAT-1 expression were subjected to analyse the diagnostic value of PCAT-1 expression (Ge et al., 2013; Shi et al., 2015; Yan et al., 2015; Qin et al., 2016; Cui et al., 2017). Table 4 shows the sensitivity and specificity of five studies related to the evaluated tumor types: ESCC, GC, HCC, and CRC. Figure 4 reveals the Forest plots of sensitivity (SEN) and specificity (SPE) of PCAT-1 expression for tumor diagnosis.
Table 4.
Summary of PCAT-1 Expression Levels as Biomarkerof Predict Prognosis in Cancers
| Study | Sample size | SE (%) | SP (%) | AUC | 95% CI | Sample |
|---|---|---|---|---|---|---|
| Ge 2013 | 58 (50)* | 78 | 54 | 0.71 | 0.62-0.76 | Tissue |
| Yan 2015 | 58 (59) | 61 | 63 | 0.66 | 0.57-0.71 | Tissue |
| Shi 2015 | 26(104) | 58 | 70 | 0.7 | 0.63-0.74 | Tissue |
| Qin 2016 | 71 (250) | 58 | 60 | 0.62 | 0.58-0.66 | Tissue |
| Cui 2017 | 87 (88) | 42 | 81 | 0.65 | 0.57-0.69 | Tissue |
, low expression (high expression)
Figure 4.
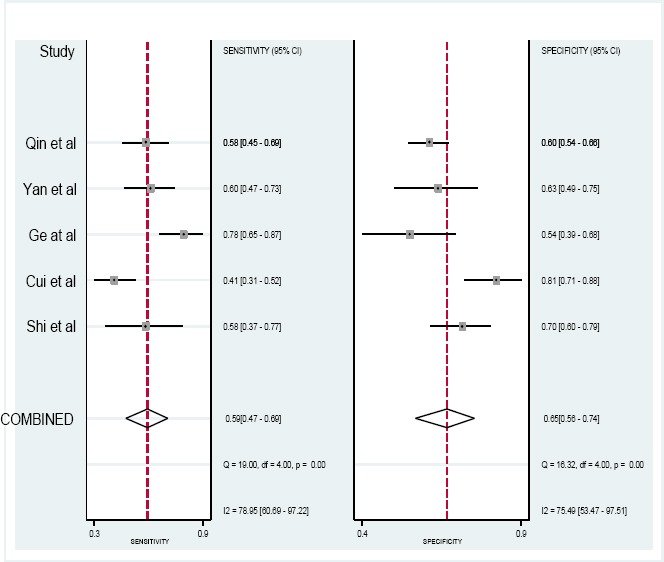
Forest Plot of Sensitivity and Specificity of PCAT-1 in Diagnosis of Cancers
Significant heterogeneity among the studies was observed with respect to both sensitivity and specificity (I2=0.81 and I2=0.79, respectively). Therefore, random-effects model was performed to summarize the diagnostic parameters. Moreover, pooled sensitivity and specificity values were 0.59 (95% CI, 0.46–0.73) and 0.66 (95%CI, 0.46– 0.73), respectively. Furthermore, the pooled area under the curve was 0.62 (95% CI: 058–0.69). Also, the area under curve (AUC) was 0.66 (95% CI 0.61–0.70), as shown in Figure 5. The ROC curve was symmetrical and the AUC was 0.66, which presents an advanced diagnostic accuracy for diagnosing of various cancers. The results reveal that lncRNA PCAT-1 have an advanced diagnostic value for esophageal squamous cell carcinoma, colorectal, prostate, hepatocellular carcinoma and gastric cancers.
Figure 5.
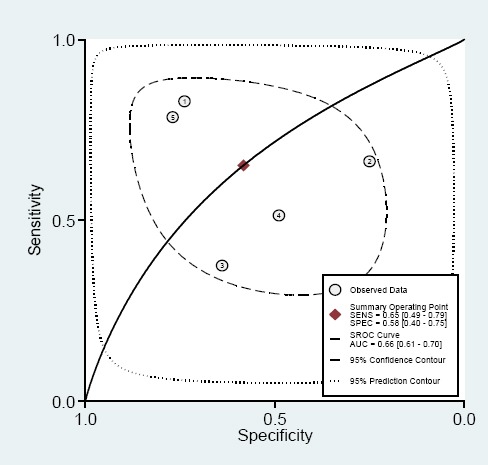
Summary Receiver Operating Characteristic Curves (SROC) for lncRNA PCAT-1 Expression Outline in the Diagnosis of Several Cancers. Every circle represents an included study.
Discussion
Aberrant expression of some lncRNAs has been demonstrated to be associated with tumor progression, as well as with the clinical outcome in various cancers (Xue et al., 2016; Qian et al., 2017; Su et al., 2017; Zhuo and Kang., 2017). Until recently, lncRNA PCAT-1 has been considered as an potential diagnostic/ prognostic biomarker in a wide variety of neoplasms, including ovarian cancer (Qian et al., 2017; Cao et al., 2014), prostate cancer (Karlsson et al., 2016; Chandra Gupta et al., 2017), colorectal cancer (Qi et al., 2013; Xu et al., 2017), non-small-cell lung cancer (NSCLC), and esophageal squamous cell carcinoma (ESCC) (Shi et al., 2015). Lnc-PCAT-1 was firstly known as a prostate-specific regulator of cell proliferation correlated with the degree of cell differentiation. Also, it was characterized as a main kind of lncRNA transcribed from the introns of known genes (Reis et al., 2004; Ren et al., 2013). The integrative analysis showed that PCAT-1, among other dysregulated lncRNAs, is a potential candidate lncRNA regulated by noncoding risk-associated SNPs in prostate cancer (Guo et al., 2016). An extensive study using RNA-Seq on prostate tissues and cells lines was completed previously (Prensner et al., 2011). They used a transcriptome assembly method to find out unannotated ncRNAs. Moreover, they recognized several dysregulated ncRNAs uncharacterized in prostate cancer (Prensner et al., 2011). Their result revealed the function of ncRNA transcriptome and confirmed dysregulation of three important target genes, BRCA2, CENPE and CENPF in prostate cancer. Altogether, the findings supported the biological role of lnc PCAT-1 as a tumor suppressor lncRNA in prostate cancer (Prensner et al., 2011). Lnc PCAT-1 was also identified as a novel prostate-specific regulator of cell proliferation and target of the Polycomb repressive complex 2 (PRC2) (Prensner et al., 2011). Furthermore, it was suggested that PCAT-1 gene may be function as a novel mechanism of “BRCA-ness” in sporadic cancers (Prensner et al., 2014).
The full texts of 5 articles among a total of 50 eligible studies were reviewed in order to meet the selection requirements for inclusion. In the systematic review and meta-analysis, it was attempted to collect all published papers assessing the prognostic significance of long non-coding RNA PCAT-1 dysregulation in several cancers. The meta-analysis was carried out on overall survival (OS) in several carcinomas, including colorectal (CRC), gastric (GC), hepatocellular carcinoma (HCC), breast, bladder, and lung cancer, extrahepatic cholangiocarcinoma, multiple myeloma (MM) and esophageal squamous cell carcinoma (ESCC). This is the first systematic review and meta-analysis in the field.
The analyses showed that the patients with high lncRNA PCAT-1 expression had evidently poorer overall survival rates than those with low PCAT-1 expression. The PCAT-1 expression level in the aforementioned malignancies was associated with metastasis, histologic grade and TNM stage. The findings revealed that PCAT-1 expression could be a potential diagnostic target in patients with colorectal (CRC), gastric, breast, bladder, lung cancer, and hepatocellular carcinoma, extrahepatic cholangio carcinoma, multiple myeloma and esophageal squamous cell carcinoma (ESCC).
This lncRNA was firstly reported to overexpress in prostate cancer and established to relate with prognosis of disease. LncRNA PCAT-1 has been demonstrated to promote cell proliferation through binding to polycomb repressive complex 2 (PCR2) as a transcriptional repressor. Based on functional experiments, lnc PCAT-1 was suggested as a potential therapeutic target for prostate cancer. Recently, the study of the expression levels of PCAT-1 in CRC tissues and matched adjacent normal tissues revealed that there is a correlation between PCAT-1 overexpression and the progression of CRC. This increased level of PCAT-1 was also associated with patients’ survival rate (Bi et al., 2017).
PCAT-1 also was shown to up-regulate in CRC tissues and correlate with overall survival and lymph node metastasis in cancer patients. Multivariate analysis demonstrated PCAT-1 as an independent prognostic factor for CRC. Moreover, related findings presented a molecular mechanism by which PCAT- 1 is implicated in CRC progression, suggesting it as a proper target for adjuvant therapy (Prensner et al., 2014).
PCAT-1 could play an important role as predictive and prognostic biomarker in some human solid tumors. However, there are some limitations to explain the results of this meta-analysis, although PCAT-1 was considered not only as potential independent predictive biomarker in survival of cancer patients. For example, there are only five studies in our study and all patients were Chiness population, it means that our results can be appropriate for Chinese population. Moreover, a significant heterogeneity in some clinicopathological characteristic was revealed. It means that there are needs to more studies with well-designed and larger-size.
Abbreviations
CRC: colorectal cancer; EMT: Epithelial-mesenchymaltransition; lincRNA, long intergenic non-coding RNA, MM: Multiple myeloma, ECC: Extrahepatic cholangiocarcinoma, PARP-1: Poly [ADP-ribose] polymerase 1, NSCLC:Non-small-cell lung carcinoma.
Conflict of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by Iran University of Medical Sciences.
References
- 1.Adams BD, Parsons Ch, Walker L, et al. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127:761–71. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 3.Bi M, Yu H, Huang B, Tang C. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in gastric cancer cells through regulating CDKN1A. Gene. 2017;626:337–43. doi: 10.1016/j.gene.2017.05.049. [DOI] [PubMed] [Google Scholar]
- 4.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;42:1–13. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. In J Cancer. 2017;140:1955–67. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 6.Cui W, Wu Y, Qu H. Up-regulation of long non-coding RNA PCAT-1 correlates with tumor progression and poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21:3021–7. [PubMed] [Google Scholar]
- 7.Evans J, Prensner J, Chen W, et al. The long noncoding RNA PCAT-1 promotes prostate cell proliferation through posttranscriptional stabilization of the cMyc oncogene. Int J Radiat Oncol Biol Phys. 2014;90:S90. [Google Scholar]
- 8.Ge X, Chen Y, Liao X, et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 9.Guo H, Ahmed M, Zhang F, et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat Genet. 2016;48:1142–50. doi: 10.1038/ng.3637. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Deng G, Liu T, et al. Long noncoding RNA PCAT-1 acts as an oncogene in osteosarcoma by reducing p21 levels. Biochem Biophys Res Commun. 2018;495:2622–9. doi: 10.1016/j.bbrc.2017.12.157. [DOI] [PubMed] [Google Scholar]
- 11.Karlsson O, Baccarelli AA. Environmental health and long non-coding RNAs. Curr Environ Health Rep. 2016;3:178–87. doi: 10.1007/s40572-016-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Cao Y, Gong X, et al. Long noncoding RNAs in head and neck cancer. Oncotarget. 2017;8:10726. doi: 10.18632/oncotarget.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Liu Y, Zhuang C, et al. Inducing cell growth arrest and apoptosis by silencing long non-coding RNA PCAT-1 in human bladder cancer. Tumor Biol. 2015;36:7685–9. doi: 10.1007/s13277-015-3490-3. [DOI] [PubMed] [Google Scholar]
- 14.Ma T, Zhou L, Xia J, et al. LncRNA PCAT-1 regulates the proliferation, metastasis and invasion of cervical cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:1907–13. doi: 10.26355/eurrev_201804_14713. [DOI] [PubMed] [Google Scholar]
- 15.Parikshak NN, Swarup V, Belgard TG, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–7. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prensner JR, Chen W, Han S, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–8. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prensner JR, Chen W, Iyer MK, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–60. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing identifies PCAT-1, a novel lincRNA implicated in prostate cancer progression. Nat Biotechnol. 2011;29:742. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi P, Xu Md, Ni Sj, et al. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian Y, Liu D, Cao S, et al. Upregulation of the long noncoding RNA UCA1 affects the proliferation, invasion, and survival of hypopharyngeal carcinoma. Mol Cancer. 2017;16:68. doi: 10.1186/s12943-017-0635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao L, Liu X, Tang Y, et al. Down regulation of the long non-coding RNA PCAT-1 induced growth arrest and apoptosis of colorectal cancer cells. Life Sci. 2017;188:37–44. doi: 10.1016/j.lfs.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Qiao L, Liu X, Tang Y, et al. Knockdown of long non-coding RNA prostate cancer-associated ncRNA transcript 1 inhibits multidrug resistance and c-Myc-dependent aggressiveness in colorectal cancer Caco-2 and HT-29 cells. Mol Cell Biochem. 2018;441:99–108. doi: 10.1007/s11010-017-3177-8. [DOI] [PubMed] [Google Scholar]
- 24.Qin HD, Liao XY, Chen YB, et al. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am J Hum Genet. 2016;98:709–727. doi: 10.1016/j.ajhg.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reis EM, Nakaya HI, Louro R, et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–92. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- 26.Ren S, Wang F, Shen J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer Care. 2013;49:2949–59. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 27.Sarrafzadeh S, Geranpayeh L, Ghafouri-Fard S. Expression analysis of long non-coding PCAT-1in breast cancer. Int J Hematol Oncol Stem Cell Res. 2017;11:185–91. [PMC free article] [PubMed] [Google Scholar]
- 28.Saus E, Brunet-Vega A, Iraola-Guzmán S, et al. Long non-coding RNAs as potential novel prognostic biomarkers in colorectal cancer. Front Genet. 2016;7:54. doi: 10.3389/fgene.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen X, Zhang Y, Wu X, et al. Upregulated lncRNA-PCAT1 is closely related to clinical diagnosis of multiple myeloma as a predictive biomarker in serum. Cancer Biomark. 2017;18:257–63. doi: 10.3233/CBM-160158. [DOI] [PubMed] [Google Scholar]
- 30.Shi Wh WuQq, Li Sq , et al. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumor Biol. 2015;36:2501–7. doi: 10.1007/s13277-014-2863-3. [DOI] [PubMed] [Google Scholar]
- 31.Su J, Zhang E, Han L, et al. Long noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis. 2017;8:e2665. doi: 10.1038/cddis.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Jin Y, Ren HX, et al. Downregulation of the long non-coding RNA TUSC7 promotes NSCLC cell proliferation and correlates with poor prognosis. Am J Transl Res. 2016;8:680–7. [PMC free article] [PubMed] [Google Scholar]
- 33.Wen J, Xu J, Sun Q, Xing C, Yin W. Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Mol Med Rep. 2016;13:4481–6. doi: 10.3892/mmr.2016.5075. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Zhao J, Zhang R. The novel long noncoding RNA TUSC7 inhibits proliferation by sponging miR-211 in colorectal cancer. Cell Physiol Biochem. 2017;41:635–44. doi: 10.1159/000457938. [DOI] [PubMed] [Google Scholar]
- 35.Xu T, Hu XX, Liu XX, et al. Association between SNPs in long non-coding RNAs and the risk of female breast cancer in a Chinese population. J Cancer. 2017;8:1162–9. doi: 10.7150/jca.18055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, Chang J, Du X, Hou J. Long non-coding RNA PCAT-1 contributes to tumorigenesis by regulating FSCN1 via miR-145-5p in prostate cancer. Biomed Pharmacother. 2017;95:1112–8. doi: 10.1016/j.biopha.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 37.Xue D, Zhou C, Lu H, et al. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumor Biol. 2016;37:16187–97. doi: 10.1007/s13277-016-5429-8. [DOI] [PubMed] [Google Scholar]
- 38.Yan TH, Yang H, Jiang JH, et al. Prognostic significance of long non-coding RNA PCAT-1 expression in human hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:4126–31. [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang D, Cao J, Zhong Q, et al. Long noncoding RNA PCAT-1 promotes invasion and metastasis via the miR-129-5p-HMGB1 signaling pathway in hepatocellular carcinoma. Biomed Pharmacother. 2017;95:1187–93. doi: 10.1016/j.biopha.2017.09.045. [DOI] [PubMed] [Google Scholar]
- 40.Zhang F, Wan M, Xu Y, et al. Long noncoding RNA PCAT1 regulates extrahepatic cholangiocarcinoma progression via the Wnt/β-catenin-signaling pathway. Biomed Pharmacother. 2017;94:55–62. doi: 10.1016/j.biopha.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q, Matsuura K, Kleiner DE, et al. Analysis of long noncoding RNA expression in hepatocellular carcinoma of different viral etiology. J Transl Med. 2016;14:328. doi: 10.1186/s12967-016-1085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao B, Hou X, Zhan H. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in non-small cell lung cancer cells. Int J Clin Exp Med. 2015;8:18482–7. [PMC free article] [PubMed] [Google Scholar]
- 43.Zhen Q, Gao LN, Wang RF, et al. LncRNA PCAT-1 promotes tumour growth and chemoresistance of oesophageal cancer to cisplatin. Cell Biochem Funct. 2018;36:27–33. doi: 10.1002/cbf.3314. [DOI] [PubMed] [Google Scholar]
- 44.Zhuo W, Kang Y. Lnc-ing ROR1-HER3 and Hippo signalling in metastasis. Nat Cell Biol. 2017;19:81–3. doi: 10.1038/ncb3467. [DOI] [PubMed] [Google Scholar]


