Abstract
Background:
In some cancer cells, the OGG1 gene is somatically mutated and highly populated. This study was conducted to examine whether OGG1 rs1052133 polymorphism is associated with the genetic background of chronic myelogenous leukaemia (CML) in Sudan.
Methods:
A total of 332 CML patients and 70 healthy controls were included in this study. Overall, the genotypes (P=0.0000) and allele (C vs. G, P=0.0007) differed considerably in the frequencies of OGG1 rs1052133 polymorphism between CML patients and controls. Our study is the first to evaluate the association of polymorphism with CML risk with OGG1 rs1052133.
Results:
A statistically significant association was observed between the genotype distribution of OGG1 rs1052133 polymorphism and CML (P=0.0000) patients. A similar result was also observed in the allele distribution (C vs. G, P=0.0007) compared with healthy controls when compared OGG1 rs1052133 genotypes with CML stages.
Results:
Genotype and allele frequencies of OGG1 rs1052133 among CML patients. A statistically significant association was observed between the genotype distribution of the OGG1 rs1052133 polymorphism and CML patients (P=0.0000). A similar result was also observed in the allele distribution (C vs. G, P=0.0007) compared with healthy controls with stages of CML in OGG1 rs1052133 genotypes.
Conclusion:
The results suggest that single nucleotide polymorphism in the gene involved in the restoration of DNA base excision (OGG1 rs1052133) can play a key role in the risk of appearance of CML. To clarify the role of OGG1 in the genetic basis of CML, further case control with larger sample sizes and fine-mapping is required.
Keywords: CML, OGG1 rs1052133, polymorphism, genotypes
Introduction
Inherited genomic variations, such as single nucleotide polymorphisms (SNPs), may also affect the genetic susceptibility to cancer (Ana Cristina et al., 2017). Identification of susceptible genes can improve understanding of leukemogenicity mechanisms and allow people at risk of growing haematological malignancies an opportunity to detect possible biomarkers and recognise disorder development, which will increase chances of survival (O’Hagan et al., 2011). Base excision repair (BER) is an essential DNA repair pathway for DNA base damage and single strand breaks caused by X-rays, oxygen radicals or alkylating agents (Frosina, 2004).
8-Oxoguanine DNA glycosylate 1 (OGG1) is a gene that is encoded by DNA. Its protein product is involved in the base oxygen repair process (BOR). The protein is responsible for the repair of 7, 8-dihydro-8-oxolamine (8-OHG). 8-OHG is a necessary lesion resulting from reactive oxygen species. Consequently, in vitro the (deficient cell lines OGG1) and in vivo were studied substantially extensively (Hirano, 2008). The OGG1 gene is somatically mutated in most cancer cells. This gene is very common among various populations. Systematic search of databases conducted by Sanjari et al. identified many studies on the relationship between breast cancer and susceptible alleles in the inheritance and population-based excision repair genes (Sanjari Moghaddam et al., 2016). Some mutated and polymorphic OGG1 protein repair activities are lower than those of the OGG1 Ser326 protein and, therefore, should be affected by human carcinogenesis (Shinmura and Yokota, 2001). OGG1 is encoded on chromosome 3p25 by the OGG1 gene. OGG1 catalyses the cleavage of the glycoside bond between the modified base and the sugar moiety, while leaving a fundamental element in DNA. Then, the product is incised. As a result, phosphodiesterase, DNA polymerase and DNA ligase are successively restored (Haixin et al., 2008; Zheng et al., 2011). Because myeloid neoplasia’s are multifactorial disorders, the study of unique gene variants and pathways can contribute to a deeper understanding of their molecular mechanisms. Therefore, we investigated the association of OGG1 rs1052133 polymorphism with genetic susceptibility to CML in Sudanese population. We performed stratified analysis to locate the most common polymorphism of OGG1 rs1052133 and to identify its association with CML based on the stage of disorder and haematological toxicity.
Materials and Methods
A total of 332 patients and 70 healthy controls were included in this study. The patients have already been identified to have CML and were either inpatients or outpatients at the Khartoum Isotope Centre in Sudan. To obtain information from all participants, informed consent was used. Venous blood samples were accumulated and stored in Ethylenediaminetetraacetic acid (EDTA) containers. XP-300 haematology analyser (Sysmex, Japan) was used to perform the complete blood count (CBC). Genomic DNA was transferred into leukocytes from peripheral blood using the general phenol-chloroform method. Based on the literature, frequent snapshots of the BER gene were acquired, including OGG1 rs1052133. SNP occurs as soon as it is genotyped in all of the samples. The PCR-CTPP approach was used to analyse genetic polymorphisms, as previously described (Hao et al., 2014). The first pairs were designed for the allele as well as the product period. Allele was based on the length of the product and primers were equal primers are based on the GenBank series (https:/www.genome.jp). We used the C allele (252 bp); F1: 5′-CAGCCCAGACCCAGTGACTC-3′ and R1: 5′-TGGCTCCTGAGCGGCGGG-3′. For the G allele (194 bp); F2: 5′-CAGTGCCCGACCGCCGCATG-3′ and R2: 5′-GGTAGTCACAGGCCCCC-3′. PCR amplification was performed using glass capillaries in a 25-μL aggregate. Using agarose gel electrophoresis, PCR products were analysed. The results of genotypes were frequently verified via random determination of 5% of the samples in DNA sequencing. All of the techniques used here are based on the ethical standards of the institutional and/or national research committee and the Helsinki Declaration of 1964 and its subsequent amendments or similar requirements. Statistical analyses were performed using SPSS (Windows v.21.0). The Hardy-Weinberg equilibrium (HWE) distribution of the OGG1 rs1052133 polymorphism genotype used the chi-squire. Odds ratios were calculated and given at 95% confidence intervals (95% CI) using an unconditional logistic regression evaluation with respect to age, gender and stage adjustment of the CML. The exact test analysis was based on OGG1 rs1052133 gene polymorphisms, as the CML and its phases have been determined by exposure variables. The p<0.05 level changed as a statistically significant consideration.
Results
Table 1 shows patient characteristics. Specifically, the mean age was 56.3 years. The patients were divided into three age groups [<40 (19.9%), 40-60 (29.5%) and >60 (50.6%) years old]. Distribution of sexes among the study groups was as follows: 204 males (76.6%) and 128 females (38.6%). There was no significant association between age and sex compared with the control groups (P= 8521, P= 0.3272). Laboratory analysis was performed for both the patients and the controls. Specifically, the Hb level, absolute neutrophil count, total lymphocyte count, platelet count and CD4 marker level were determined. The results showed a statistically significant decrease in all levels relative to the control groups (P<0.0001). The variation in genotype distribution between the CML patients and the controls, and the associations between CML and risk allele of the OGG1 rs1052133 polymorphism were evaluated by accounting for age and gender. The genotype and allele frequencies of OGG1 rs1052133 among the CML patients are shown in Table 2. A statistically significant association was found between the genotype distribution of the OGG1 rs1052133 polymorphism and the CML patients (P=0.0000). A statistically significant association was also observed in the allele distribution (C vs. G, P=0.0007) compared with healthy controls. When determining the association of OGG1 rs1052133 genotypes with stages of CML, the results showed that the OGG1 rs1052133 genotype was significantly correlated with the accelerated (p=0.0204) and acute (P=0.0000) CML stages. However, as shown in Table 3, there were no correlations detected in chronic stage of CML (p = 0.1873). The frequencies of Febrile neutropenia associated with CML stages were determined for different stages of the disease (Figure 1). Neutropenia increased with increase of the disease stage. At OGG1 rs1052133 (Figure 2), CD4 concentrations were increased in GG compared with CG and CC genotypes.
Table 1.
General Characteristics of CML Patients and Control
| Mean ± SD | P. value☐ | ||
|---|---|---|---|
| Control (n = 70) | Patients (n =332) | ||
| Age/years | |||
| < 40 n (%) | 16 (22.8) | 66 (19.9) | |
| 40 -60 n (%) | 20 (28.6) | 98 (29.5) | 0.8521 |
| > 60 n (%) | 34 (48.6) | 168 (50.6) | |
| Gender | |||
| Male n (%) | 38 (54.3) | 204 (61.4) | |
| Female n (%) | 32 (45.7) | 128 (38.6) | 0.3272 |
| Laboratory analysis | |||
| Hb g/dl | 12.3±2.1 | 9.3±3.3 | 0.0001 |
| Neutrophils c/µ | 4.1±3.6 | 1.9±2.3 | 0.0001 |
| Lymphocytes c/µ | 2.6±4.2 | 5.6±2.1 | 0.0001 |
| Platelet c/µ | 280.2±3.5 | 760±3.1 | 0.0001 |
| CD4 % | 37.3±1.5 | 19.7±1.9 | 0.0001 |
P < 0.05. is referred as statistically significant using t. independent test and fisher exact test
Table 2.
The Frequency of OGG1 rs1052133 Genotypes and Alleles in CML Patients
| Frequency n (%) | |||
|---|---|---|---|
| Genotype | Control, n = 70 | Patients, n=332 | P. value* |
| Homozygous | 264 (79.5) | ||
| CC | 40 (51.2) | ||
| Heterozygous | 50 (15.1) | ||
| CG | 26 (37.1) | 0 | |
| Wild type | 18 (5.4) | ||
| GG | 4 (5.7) | ||
| Allele | |||
| C | 106(75.7) | 589(87.0) | 0.0007 |
| G | 34(24.3) | 86(13.0) | |
, Fisher exact test was used to determine the p. value (p <0.05 is significant) after adjusted for age and gender of CML.
Table 3.
The Genotypes Distribution and Odds Ratios among CML Phases and Controls
| Frequency n (%) | |||
|---|---|---|---|
| Genotype | Patients | Control | P. value* |
| Chronic Stage (n =34) | |||
| CC | 14 (41.2) | 40 (51.2) | |
| CG | 19 (55.9) | 26 (37.1) | 0.1873 |
| GG | 1 (2.9) | 4 (5.7) | |
| Accelerated Stage (n = 68) | |||
| CC | 51 (75.0) | 40 (51.2) | |
| CG | 11 (16.2) | 26 (37.1) | 0.0204 |
| GG | 6 (8.8) | 4 (5.7) | |
| Acute Stage (n =230) | |||
| CC | 199 (86.5) | 40 (51.2) | |
| CG | 20 (8.6) | 26 (37.1) | 0 |
| GG | 11 (4.9) | 4 (5.7) | |
, chi-square analysis was performed to determine the p. value (p <0.05 is significant) after adjusted for age and family history of CML.
Figure 1.
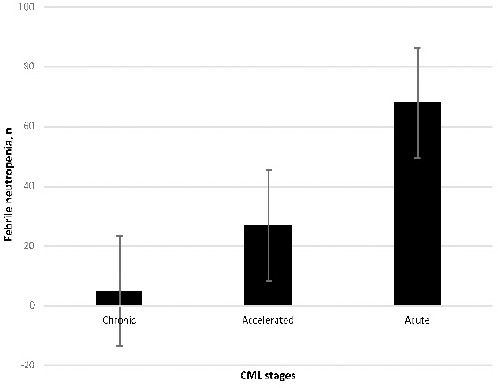
The Frequency of Febrile Neutropenia Associated to the Stages of CML (The neutropenia was increased with the stage of the disease, It was more in acute stage).
Figure 2.
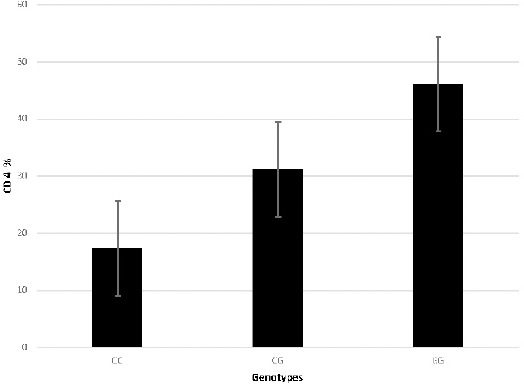
The CD 4 Concentration levels Associated to the Genotype’s Frequencies in CML Patients (CD4 concentrations were elevated in GG compared to CG and CC genotypes at OGG1 rs1052133).
Discussion
Compared to previous studies (Ahmed et al., 2009; Farzana et al., 2015), the results of the present study (i.e., frequencies vs. sex) showed that males were more affected than females. Most of our patients were over 60 years of age and were in the blast phase of CML (acute stage) compared to Farzana’s and other studies (Farzana et al., 2015). The BER pathway eliminates many varieties of basic damage through several coordinated sequential reactions, which recognize and repair damage that results from reactive oxygen species, hydroxylation and various cellular processes (Bensu et al., 2014; Maynard et al., 2009). Therefore, genetic polymorphism in BER genes can affect variations in the DNA repair capacity of males and females. In addition, this can be linked to the increased risk of CML. When we assessed the relationship between sequence versions in the BER gene (OGG1 rs1052133), our results confirmed that these genotypes were significantly correlated with CML patients. This indicates the influence of the OGG1 variant, which is consistent with previous studies on bladder and breast cancer (Hao et al., 2014; Bensu et al., 2014; . Maynard et al., 2009). Our results indicate that OGG1 rs1052133 genotypes (CC and CG) promoted the development of CML, which is in contrast to the GG genotype. This suggests that CC genotypes are significantly correlated with the risk of CML. These findings indicate that specific genotypes within a single repair pathway are factors that affect the risk of CML. Because SNP rs1052133 significantly interacts with numerous variants of proteins with a slightly accelerated or reduced useful activity (Zheng et al., 2011; Mohammed et al., 2017), DNA repair activity may be also affected, which increases the risk of CML. The advantage of our study is that we investigated OGG1 rs1052133 polymorphism among the patients with CML in Sudan. In addition, this work is the first to examine the role of polymorphism in OGG1 rs1052133. Moreover, the patients and the controls were assessed based on their age and gender. The limitation of our study is that the study sample was relatively modest. Therefore, the output of SNP effects needs to be further validated in large population samples.
In conclusion, our results indicate that single nucleotide polymorphism in the gene involved in DNA base excision (OGG1 rs1052133) might play a crucial role in CML development, and could play an important role in both an accelerated risk of CML and patient survival. The variants of this gene influence the predisposition to strengthen CML and worsen the prognosis of patients with DNA damage with this neoplasm. To confirm our findings and the possible relationship between DNA repair (OGG1 rs1052133 gene polymorphism) and CML risk, larger population studies involving different BER genes are needed.
References
- 1.Ahmed R, Naqi N, Hussain I, et al. Presentating phases of chronic myeloid leukaemia. See comment in PubMed Commons below. J Coll Physicians Surg Pak. 2009;19:469–72. [PubMed] [Google Scholar]
- 2.Ana Cristina G, Raquel A, Ines B, et al. Genetic variants involved in oxidative stress, base excision repair, DNA methylation, and folate metabolism pathways influence myeloid neoplasias susceptibility and prognosis. Mol Carcinog. 2017;56:130–48. doi: 10.1002/mc.22478. [DOI] [PubMed] [Google Scholar]
- 3.Bensu K, Ays¸e Bas¸ak E, Erdem C, et al. Could 8-oxoguanine DNA glycosylase 1 Ser326Cys polymorphism be a biomarker of susceptibility in cancer? Toxicol Ind Health. 2014;30:814–25. doi: 10.1177/0748233712463777. [DOI] [PubMed] [Google Scholar]
- 4.Farzana C, Riaz Q, Mehrab K, et al. Clinico hematological profile and phase distribution of chronic myeloid leukemia. Biol Med (Aligarh) 2015;7:100025. [Google Scholar]
- 5.Frosina G. Commentary: DNA base excision repair defects in human pathologies. Free Radical Res. 2004;38:1037–54. doi: 10.1080/10715760400011445. [DOI] [PubMed] [Google Scholar]
- 6.Haixin L, Xishan H, Wei Z, Qingyi W, Kexin C. The hOGG1 Ser326Cys polymorphism and lung cancer risk: A meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;2008:17. doi: 10.1158/1055-9965.EPI-08-0001. [DOI] [PubMed] [Google Scholar]
- 7.Hao L, Zheng L, Yi Q, et al. Single nucleotide polymorphisms of DNA base-excision repair genes (APE1, OGG1 and XRCC1) associated with breast cancer risk in a Chinese population. Asian Pac J Cancer Prev. 2014;15:1133–40. doi: 10.7314/apjcp.2014.15.3.1133. [DOI] [PubMed] [Google Scholar]
- 8.Hill JW, Evans MK. A novel R229Q OGG1 polymorphism results in a thermolabile enzyme that sensitizes KG-1 leukaemia cells to DNA damaging agents. Cancer Detect Prev. 2007;31:237–43. doi: 10.1016/j.cdp.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano T. Repair system of 7, 8-dihydro-8-oxoguanine as a defense line against carcinogenesis. J Radiat Res (Tokyo) 2009;49:329–40. doi: 10.1269/jrr.08049. [DOI] [PubMed] [Google Scholar]
- 10.Hu JJ, Smith TR, Miller MS, et al. Genetic regulation of ionizing radiation sensitivity and breast cancer risk. Environ Mol Mutagen. 2002;39:208e215. doi: 10.1002/em.10058. [DOI] [PubMed] [Google Scholar]
- 11.Maynard S, Schurman SH, Harboe C, et al. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2e10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohammed A, Akbar A, Jilani S, et al. The hOGG1 Ser326Cys gene polymorphism and breast cancer risk in Saudi population. Pathol Oncol Res. 2017;23:525–35. doi: 10.1007/s12253-016-0146-6. [DOI] [PubMed] [Google Scholar]
- 13.O'Hagan HM, Wang W, Sen S, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and Polycomb Members to promoter CpG islands. Cancer Cell. 2011;20:606–19. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petermann E, Keil C, Oei SL. Roles of DNA ligase III and XRCC1 in regulating the switch between short patch and long patch BER. DNA Repair (Amst) 2006;5:544e555. doi: 10.1016/j.dnarep.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Sanjari Moghaddam A, Nazarzadeh M, Noroozi R, Darvish H, Mosavi Jarrahi A. XRCC1 and OGG1 gene polymorphisms and breast cancer: a systematic review of literature. Iran J Cancer Prev. 2016;9:e3467. doi: 10.17795/ijcp-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen M, Hung RJ, Brennan P, et al. Polymorphisms of the DNA repair genes XRCC1, XRCC3, XPD, interaction with environmental exposures, and bladder cancer risk in a case-control study in Northern Italy. Cancer Epidemiol Biomarkers Prev. 2003;12:1234e1240. [PubMed] [Google Scholar]
- 17.Shinmura K, Yokota J. The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis. Antioxid Redox Signal. 2001;3:597–609. doi: 10.1089/15230860152542952. [DOI] [PubMed] [Google Scholar]
- 18.Smith TR, Levine EA, Perrier ND, et al. DNA repair polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:1200e1204. [PubMed] [Google Scholar]
- 19.Yu P, Zheng L, Shiheng Z, Yanli X, et al. Association of DNA base excision repair genes (OGG1, APE1 and XRCC1) polymorphisms with outcome to platinum-based chemotherapy in advanced non-small-cell lung cancer patients. Int J Cancer. 2014;135:2687–96. doi: 10.1002/ijc.28892. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L, Wei G, Meng-xia L, et al. Genetic polymorphism of DNA base-excision repair Genes (APE1 OGG1 and XRCC1) and their correlation with risk of lung cancer in a Chinese population. Arch Med Res. 2011;42:226–34. doi: 10.1016/j.arcmed.2011.04.005. [DOI] [PubMed] [Google Scholar]


