Figure EV2. The p97–ATX3 complex regulates RNF8 turnover at sites of DNA damage.
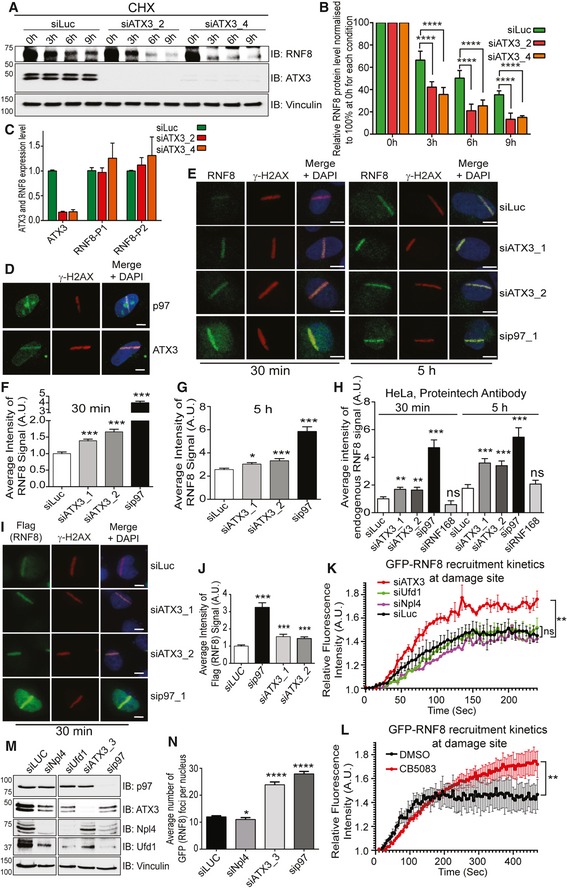
- Western blot analysis of CHX chase kinetics showing accelerated RNF8 degradation in soluble fraction (cytoplasm + nucleoplasm) of siATX3‐depleted HeLa cells.
- Graph represents the quantifications of (A) (****P < 0.0001; two‐way ANOVA, n = 3, mean + SEM).
- ATX3 and RNF8 mRNA expression level analysed by quantitative PCR after indicated siRNA treatment in U2OS cells. The experiment was performed in quadruplet (n = 1) and columns represent the mean + SEM.
- Representative IF micrographs of U2OS cells showing recruitment of p97 or ATX3 to UV‐A micro‐laser‐induced DNA damage tracks. Scale bar: 10 μm.
- Representative IF images showing the UV‐A micro‐laser‐induced DNA damage tracks in U2OS cells. Endogenous RNF8 and γ‐H2AX signal at damage tracks after 30 min and 5 h of damage induction under indicated siRNA‐depleted conditions. Scale bar: 10 μm.
- Quantification of (E) at 30 min time point. Graph represents the average intensity of the RNF8 signal (***P < 0.001; unpaired t‐test, n = 3, mean + SEM, in average, at least 70 nuclei per condition and experiment).
- Quantification of (E) at 5‐h time point. Graph represents the average intensity of the RNF8 signal (*P < 0.05, ***P < 0.001; unpaired t‐test, n = 3, mean + SEM, in average, at least 70 nuclei per condition and experiment).
- Quantification of endogenous RNF8 signal intensity in HeLa cells at UV‐A micro‐laser‐induced DNA damage tracks 30 min and 5 h after damage induction under indicated siRNA‐depleted conditions. A second, commercially available (Proteintech) RNF8 antibody was used. Graph represents the average intensity of RNF8 signal (ns P > 0.05, **P < 0.01, ***P < 0.001; unpaired t‐test, n = 1, mean + SEM, more than 50 nuclei were analysed per condition and experiment).
- Representative IF micrographs of HeLa cells showing Flag‐RNF8 signal intensity at UV‐A micro‐laser‐induced DNA damage tracks 30 min after damage induction under indicated siRNA‐depleted conditions. Scale bar: 10 μm.
- Quantification of (I). Graph represents the average intensity of RNF8 signal (***P < 0.001; unpaired t‐test, n = 1, mean + SEM, more than 50 nuclei were analysed per condition and experiment).
- Graph represents the recruitment kinetics of GFP‐RNF8 at sites of two‐photon laser‐induced DNA damage spot in living U2OS cells under indicated conditions (ns P > 0.05, **P < 0.01; unpaired t‐test on area under curve, n = 2, for siNpl4 n = 1, mean + SEM, in average, at least five nuclei per condition and experiment).
- Graph represents the recruitment kinetics of GFP‐RNF8 at sites of two‐photon laser‐induced DNA damage spot in living U2OS cells under indicated conditions (**P < 0.01; unpaired t‐test on area under curve, n = 3, mean + SEM, in average, at least five nuclei per condition and experiment).
- Western blot analysis showing depletion efficiency of indicated siRNAs in U2OS cells.
- Graph represents the quantification of GFP (RNF8) foci in fixed U2OS cells after 30 min of IR (2 Gy) treatment, under indicated conditions (*P < 0.05, ****P < 0.0001; unpaired t‐test, n = 2, +SEM).
Source data are available online for this figure.
