Figure EV4. Monomerisation allows ATP to bind to FICD in a mode conducive to BiP AMPylation.
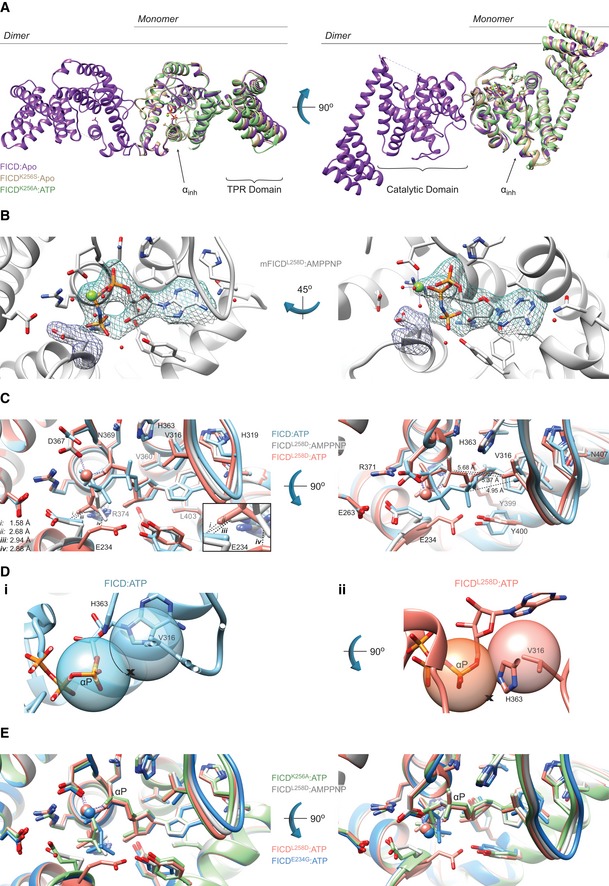
- Mutation of the dimer relay residue Lys256 does not result in large conformational changes in FICD. Shown is the alignment (residues 213–407) of the molecules in the asymmetric unit. Structures are coloured as indicated. Glu234, ATP and Mg (where applicable) are shown as sticks. The inhibitory alpha helix (αinh) and gross domain architecture is annotated. The FICD:Apo structure is from PDB: 4U04.
- Electron density of both MgAMPPNP and the inhibitory Glu234, from monomeric FICDL258D co‐crystallised with MgAMPPNP. Unbiased polder (OMIT) maps are shown in blue and purple meshes, contoured at 3.0 and 5.0σ, respectively. All residues and water molecules interacting with MgAMPPNP are shown as sticks and coloured by heteroatom. Mg2+ coordination complex pseudo‐bonds are show in purple dashed lines.
- Unlike wild‐type FICD, monomeric FICDL258D binds ATP and ATP analogues in an AMPylation‐competent conformation. The indicated structures and distances are shown as in Fig 5C, with ATP interacting residues shown as sticks and annotated. The position of the α‐phosphate relative to Val316 in the FICD:ATP structure (see distances in right‐hand side panel) would preclude in‐line nucleophilic attack (see D, E). The inset is a blow‐up displaying distances (i–iv) between the γ‐phosphates and Glu234 residues. A potentially significant difference in the Glu234 position between the FICDL258D:MgAMPPNP and FICD:ATP structures is apparent: hypothetical distance (ii) (2.68 Å, between Glu234 of FICD:ATP and AMPPNP γ‐phosphate of FICDL258D) is less favourable than the observed distance (iii) (2.94 Å, between the AMPPNP γ‐phosphate and Glu234 of FICDL258D). Note, His363 of FICD:ATP is in a non‐optimal flip state to facilitate general base catalysis (see Fig 5B).
- (i) The mode of ATP binding in wild‐type dimeric FICD sterically occludes the nucleophilic attack required for AMPylation. Shown are semi‐opaque 3 Å centroids centred on Pα and Val316 (Cγ1). The putative BiP Thr518 nucleophile (depicted by the cross) is positioned in‐line with the scissile phosphoanhydride (parallel to the plane of the paper) and 3 Å from Pα. This nucleophile position lies within the Val316 centroid (indicating a steric clash). For clarity, the FICD:ATP structure is overlaid with a thin slice of the FICD:ATP structure in the plane of the Pα‐O3α bond. (ii) In the monomeric AMPylation‐competent FICDL258D:ATP structure, the nucleophile lies outside the Val316 centroid in proximity to His363 (the general base).
- The ATP α‐phosphates of monomer or dimer relay mutants are in the same position as that competently bound to the AMPylation unrestrained dimeric FICDE234G. Shown are all AMPylation‐competent MgATP structures overlaid as in (C) and Fig 5C. The dimeric FICDE234G:MgATP (dark blue, PDB: 4U07) is also included as a reference for an active AMPylating enzyme.
