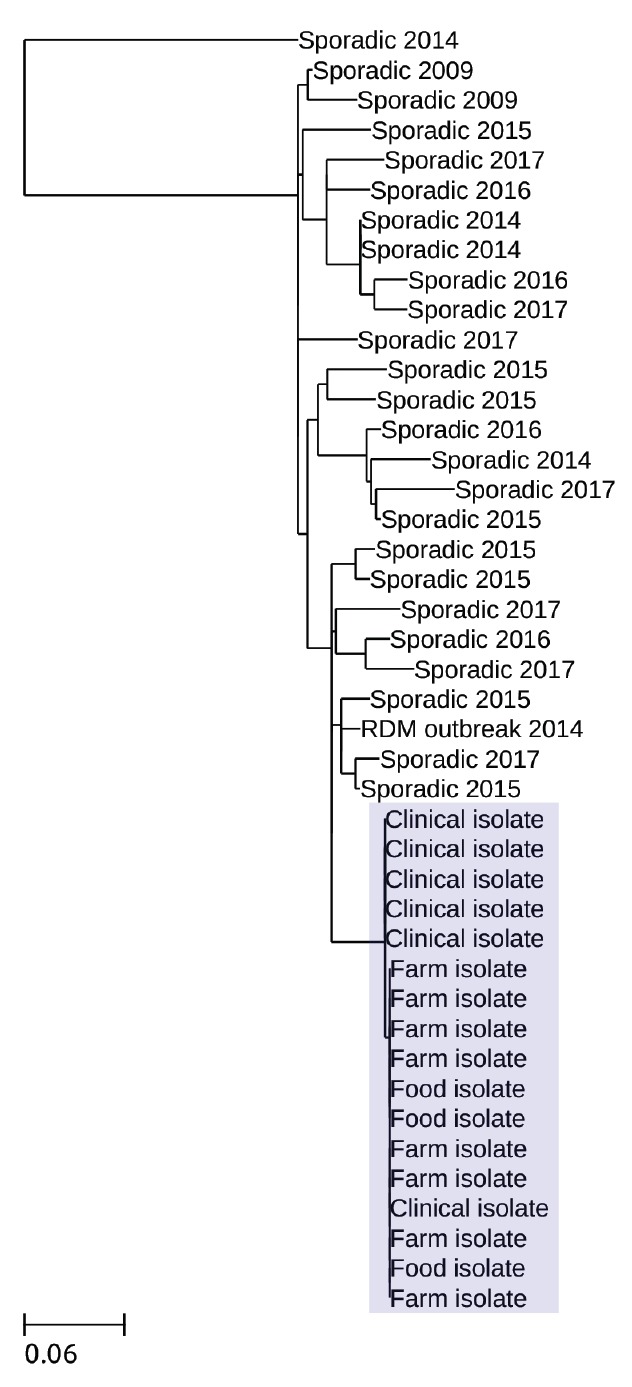
Figure 3.
Phylogeny of the clinical, food and farm isolates linked to the STEC O157:H7 outbreak, England, 2017
STEC: Shiga toxin-producing Escherichia coli.
Isolates linked to the outbreak are highlighted. The clinical isolates were from the six confirmed cases. Sequences from other closely related sporadic isolates (sporadic isolates defined here as not linked to the outbreak) within the 25 single nucleotide polymorphism (SNP) single linkage cluster available in the Gastrointestinal Bacteria Reference Unit archive are shown for context, including an isolate from an outbreak associated with raw drinking milk in 2014 in the south-west of England .Quality trimmed Illumina reads were mapped to the STEC O157 reference genome Sakai (Genbank accession number: BA000007) using BWA-MEM. SNPs were identified using GATK2 in unified genotyper mode. Core genome positions that had a high quality SNP (> 90% consensus, minimum depth 10×, MQ ≥ 30) in at least one isolate were extracted. SNP positions that were present in at least 80% of isolates were used to derive maximum likelihood phylogenies with RaxML using the GTRCAT model with 1,000 iterations.