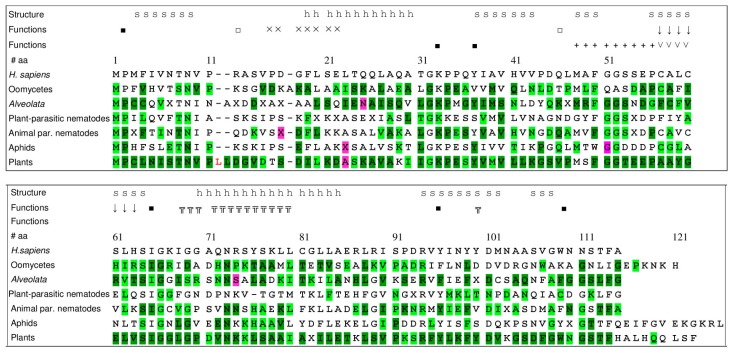
Figure 6.
Conservation of amino acid sites and motifs in consensus MIF sequences. Alignment (amino acid one letter code) of human MIF sequences and consensus sequences of oomycetes, Alveolata, plant-parasitic nematodes, animal-parasitic nematodes, aphids and plants. Amino acid numbers are given above the human sequence. Hyphens (-) indicate a gap in the respective sequence. Consensus sequences, obtained using Jalview version 2.9, show the most frequent amino acids at each position, or an X when the frequency of the most prominent amino acid is below 40%. Sites under purifying or diversifying selection are colored as follow: pink = purifying selection predicted by 4 to 5 selection algorithms; light green = purifying selection predicted by two to three selection algorithms; dark green = purifying selection predicted by four to five selection algorithms. α helices and β sheets of human MIF are indicated by the letters ‘h’ and ‘s’, respectively. Amino acids shown to determine specific functions or activities in human MIF are specified by symbols above the sequence. They are involved in the tautomerase activity (■ symbol), oxidoreductase activity (CXXC motif; ˅ symbol), binding to the CXCR4 receptor (╦ symbol), binding to the CXCR2 receptor (pseudo ELR motif; □ symbol), interaction between MIF and the CXCR2 receptor (N like loop pattern; + symbol), nuclease activity (PE/DxxxxE motif; x symbol) and DNA damage response protein. (CxxCxxHx(n) zinc finger domain; ↓ symbol).