Abstract
Hepatoblastoma (HB) is the most common form of primary liver malignancy found in pediatric populations. HB is considered to be clonal and arises from hepatoblasts, or embryonic liver progenitor cells. These less differentiated tumor-initiating progenitor cells, or cancer stem cells (CSCs), may contribute to tumor recurrence and resistance to therapies, and have high metastatic abilities. Phenotypic heterogeneity, undesired genetic and epigenetic alterations, and dysregulated signaling pathways provide CSCs with a survival advantage over current therapies. The molecular and cellular basis of HB and the mechanism of CSC induction are not fully understood. The Wnt/beta-catenin pathway is one of the major developmental pathways and is believed to play an important role in the pathogenesis of HB and CSC formation. This review summarizes the cellular and molecular characteristics of HB with a specific emphasis on CSCs and Wnt/beta-catenin signaling.
Keywords: hepatoblastoma, cancer stem cells, pediatric liver cancer, Wnt signaling, beta-catenin
1. Introduction
Hepatoblastoma (HB) is the primary liver cancer found in children generally under three years of age [1]. The term “hepatoblastoma” suggests that these tumors may originate from hepatoblasts, or embryonic liver progenitor cells. HB subtypes are identified using histological subtyping based on the level of cell differentiation. HB tumor aggressiveness is often associated with the stem cell phenotype. Therefore, identifying the mechanisms by which normal stem/progenitor cells are transformed into cancer stem cells is critical for elucidating the molecular and cellular basis of HB. Though a rare type of cancer, the annual incidence of HB has increased in recent years, accounting for nearly 80% of all malignant liver tumors in childhood and comprising about 1% of all pediatric cancers [2,3]. Although the cause of HB is unknown, there are several risk factors associated with its occurrence. These include Beckwith–Wiedemann syndrome, familial adenomatous polyposis, premature birth, low birth weight, conception after infertility treatment, maternal smoking, and a higher maternal pre-pregnancy body mass index (BMI of 25–29) [4,5,6]. Children who are exposed to hepatitis B infection at an early age or suffer from a chronic cholestatic liver disease, such as biliary atresia [7], are also at a higher risk of developing HB. Treatment options vary depending on factors such as tumor stage and metastatic nature. A complete surgical resection and chemotherapy constitute the only primary therapy for HB. Cisplatin-based chemotherapy has become an effective therapy for patients with unresectable tumors and has increased the survival rate of HB patients [8]. Despite the clinical advancements and the high survival rate, liver transplantation is the only treatment recommended in cases of advanced hepatoblastoma in close proximity to vascular structures [1]. Although chemotherapeutics have greatly increased the survival rate, detailed studies are still needed to optimize therapies for patients at high risk of distantly metastasizing and recurrent HB tumors [2,8,9,10].
2. Molecular Signatures Associated with HB
HB and HCC (hepatocellular carcinoma) are common liver malignancies that affect different age groups. Even though there are similarities in their tumorigenic characteristics, recent studies have shown that HB differs from HCC not only in its origin, but also in various molecular signatures associated with each malignancy [11]. Global gene expression analysis has demonstrated that genes such as insulin-like growth factor 2 (IGF-2), delta-like 1 homolog (DLK1), transforming growth factor beta 1 (TGF beta-1), mitogen-inducible gene 6 (MIG-6), and non-coding RNA MALAT1 are induced in HB but not in HCC [11]. On the other hand, interferon gamma-inducible protein 27, galectin, ubiquitin 2, and alpha-1 androgen receptor are downregulated in HB but not in HCC [11]. The FXR (farnesoid X receptor)–gankyrin signaling axis is another signaling pathway found to be activated in HB, along with a reduction of tumor-suppressing RNA-binding protein CUGBP1 [12].
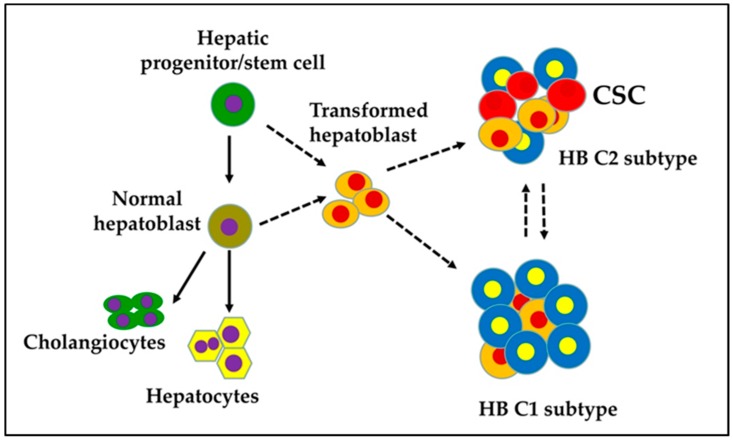
The presence of various fetal and embryonic liver progenitor markers strengthens the hypothesis that HB originates from transformed hepatoblasts. HB tumors are categorized into two distinct subtypes: C1 and C2 [13]. The C1 subtype recapitulates fetal liver histological features and expresses markers of differentiated hepatocytes. The C2 subtype resembles earlier stages of liver development with an embryonal histology and is characterized by the expression of markers of hepatic progenitor/stem cells (Figure 1). Sequencing studies have shown that HB is one of the pediatric cancers with the lowest rates of somatic mutations. The CTNNB1 gene, which encodes beta-catenin, is the most frequently mutating gene in exon 3 in HB; consequently, an adenomatous polyposis coli (APC) germline mutation in HB has been associated with a familial adenomatous polyposis history. Genomic analysis has revealed a CTNNB1 mutation rate of ≈68% in HB [14,15,16,17,18,19,20,21,22]. This indicates that Wnt/beta-catenin signaling plays a pivotal role in the malignant transformation of hepatoblasts and HB pathogenesis.
Figure 1.
Proposed cellular origin of cancer stem cells (CSC) in a hepatoblastoma (HB).
Overexpression of genes, such as AFP, TACSTD1, DLG7, CDC2, BUB1, AURKB, IGF2, DLK1, PEG3, PEG10, BEX1, MEG3, NDN, BIRC5, NPM1, and HDAC2, and the activation of Myc signaling have been associated with the HB C2 subtype, which has embryonal characteristics and is enriched with CSCs [13]. On the other hand, C1 subtype HB tumors are characterized by the overexpression of genes such as GLUL, RHBG, CYP2E1, and CYP1A1. It is also important to note that different sets of Wnt/beta-catenin-targeted genes are overexpressed in the two HB subtypes [13]. The HB subtypes are also distinguished by the differential expression of metabolic genes such as glucose transporter 3 (GLUT3), lactate dehydrogenase B (LDHB), and glucose-6-phosphate (G6PC), demonstrating distinct glycolytic events in these tumors [23]. Another gene affected by recurrent mutations in HB cases is NFE2L2 (also known as NRF2), which encodes a transcription factor involved in the antioxidant response pathway [14]. NRF2 mutations are correlated with the activation of nicotinamide adenine dinucleotide phosphate dehydrogenase quinone 1 (NQO1), which has high expression levels in HB and has been found to be associated with invasive and metastatic C2 subtype HB tumors [14]. Comerford et al. developed an interesting mouse model of HB by overexpressing Myc and mutant beta-catenin, wherein mice preferentially developed HB in an NRF2-dependent mechanism [24]. This mouse model mimics molecular characteristics of human HB and demonstrates the synergistical functional role of Myc and beta-catenin, along with an important role of antioxidant pathways, in HB development.
Additionally, cytoplasmic activation/proliferation-associated protein-2 (CAPRIN2), a known oncogene, and tumor suppressors, such as speckle-type POZ protein (SPOP), olfactory receptor-511 (OR5I1), and cell division cycle 20B (CDC20B), have been found to influence HB cell growth. CAPRIN2 is known to activate Wnt signaling via low-density lipoprotein receptor-related protein (LRP) 5/6 phosphorylation [25], and gain-of-function mutations in CAPRIN2 may enhance Wnt/beta-catenin signaling [26]. This study also reported novel mutations affecting components of ubiquitin ligase complexes, including SPOP, (kelch-like 22 KLHL22), transient receptor potential cation channel subfamily C member 4 associated protein (TRPC4AP), and ring finger protein 169 (RNF169) [26]. Studies have found that dephosphorylation of tumor suppressor protein CCAAT/enhancer-binding protein alpha (C/EBP alpha) creates pre-neoplastic foci with CSCs that gives rise to HCC and aggressive forms of HB [27]. These biomarkers can complement existing histopathological methodologies and may aid in a clinical diagnosis that requires distinguishing between aggressive forms of HB.
3. Cellular Origin of HB: Transformed Hepatoblasts
HB differs in its developmental origin from HCC in adults. HB occurs in the very early stages of life, while HCC frequently develops on a background of chronic liver diseases. Studies have demonstrated that HB tumors consist of heterogenous populations of stem/progenitor cells [10,13,28]. Therefore, HB is believed to originate from hepatoblasts that undergo undesired genetic/epigenetic aberrations during development (Figure 1). Stemness provides cells with a strong driving force for uncontrolled growth and survival, which may also be associated with metastasis, drug resistance, and tumor recurrence. CSCs possess a high level of plasticity and might undergo undesired genetic, phenotypic, and epigenetic changes in response to the tumor microenvironment, which increase their resistance to therapies and their ability to metastasize to other organs. Because of the phenotypic/genetic heterogeneity, there is no common strategy for targeting CSCs. Hence, CSCs represent the critical subset of cells within the tumor mass in perpetuating the tumor and need to be targeted effectively to prevent HB recurrence and metastasis [2].
CSCs express numerous progenitor/stem cell-specific genes and share many characteristics with normal stem cells. HB tumors have been found to express progenitor markers, such as clusters of differentiation 133 (CD133), EpCAM (epithelial cell adhesion molecule), CD44, CD24, CD90, oval cell antigen-6 (OV6), and aldehyde dehydrogenase1 (ALDH1) [29,30]. CD133, one of the widely studied stem cell markers, is a transmembrane glycoprotein that is significantly upregulated in human liver cancer, as well as in animal models of liver cancer [30,31,32,33,34]. CD133 is also expressed in hepatoblasts, as well as in cholestasis-induced epithelial–mesenchymal liver progenitor cells [35,36]. The CD133 expression profile has been investigated in several clinical studies. Increased CD133 expression levels in human samples have been correlated with tumor stages and alpha-fetoprotein levels, and are characterized by overall lower survival and prognosis and higher recurrence rates [31,37,38]. The CD133-positive tumor cells have exhibited a significantly higher colony-forming efficiency and proliferation ability in vitro and tumorigenicity in xenograft models [34]. The expression of genes involved in self-renewal pathways is markedly higher in CD133-positive cells, and studies have shown that these cells are more resistant to conventional chemotherapy [31,34]. CD133 has been found to be induced by Wnt/beta-catenin signaling in tumor-initiating cells and hepatoblastoma cells [39]. CD133-positivity has been shown to increase chemoresistance via preferential activation of the Akt/PKB and Bcl-2 survival pathways [40]. A recent study found that PIM (proviral integration site for Moloney murine leukemia) kinase promoted stemness via the induction of CD133 in HB cells. PIM kinase inhibition reduced CD133 expression and arrested the growth of hepatoblastoma tumors with CD133-enriched HB cells [41].
EpCAM is another potential CSC marker in the liver. In a normal liver, EpCAM is expressed in the bile duct cells at low levels. EpCAM is a transmembrane protein induced by Wnt/beta-catenin signaling and expressed in a variety of progenitor and stem cells [42], including hepatoblasts and adult hepatic progenitor cells [43,44,45]. EpCAM-positive CSCs have been found to be more tumorigenic and invasive compared to EpCAM-negative cells. Cleavage of EpCAM by tumor necrosis factor-alpha converting enzyme (TACE) or a disintegrin and metalloproteinase domain- containing protein 17 (ADAM17)) and a gamma-secretase complex containing presenilin 2 (PS-2) result in the release of the EpEX domain into the extracellular space and intracellular EpICD domain into the cytoplasm. EpICD directly interacts with beta-catenin and lymphoid enhancer factor (LEF) [46]. The overexpression of EpCAM in hepatic malignancies is associated with a poor survival rate [47]. Due to its high incidence in tumors, EpCAM has been one of the CSC targets in clinical studies, and specific anti-EpCAM antibodies have been tested in cancer patients [48].
CD90 and CD44 have also emerged as markers of liver CSCs. Yang et al. found that the expression of CD90 correlated with tumorigenic potential in liver cancer cell lines. The presence of CD90-positive and CD44-positive cells was found to correlate with tumor metastasis. Blockade of CD44 by a neutralizing antibody induced apoptosis of CD90-positive cells and prevented tumor formation in mice [49].
4. Epithelial–Mesenchymal Transition in HB
Epithelial–mesenchymal transition (EMT) is a cellular reprograming process whereby epithelial cells undergo transformation into cells with mesenchymal characteristics. In EMT-activated cells, intercellular junction proteins, such as E-cadherin, are downregulated, while mesenchymal-related proteins, such as N-cadherin, fibronectin, and vimentin, are upregulated. When primary tumor cells undergo EMT, polarized cells become highly migratory and are capable of invasion, leading to distant metastatic colonization [50,51]. Several mechanisms have been linked to EMT in HB. Cancer cells are known to acquire stem-like characteristics through EMT [51,52]. Periostin (POSTN)-induced hepatoblastoma cells undergo EMT, mediated by the mitogen activated protein kinase (MAPK)/extracellular signal regulated kinase (ERK) pathway [53]. DNA damage-regulated autophagy modulator 1 (DRAM1) has been reported to be associated with EMT, migration, and invasion in HepG2, an HB cell line [54]. The long non-coding RNA OIP5-AS1-miR-186a-5p-ZEB1 signaling axis has also been found to regulate EMT in HB [55]. Thymosin beta-4, a known G-actin sequestering factor, modulates the dynamic changes in the cytoskeleton, and has been shown to promote EMT in HB cells [56].
5. Wnt/Beta-Catenin Signaling in HB
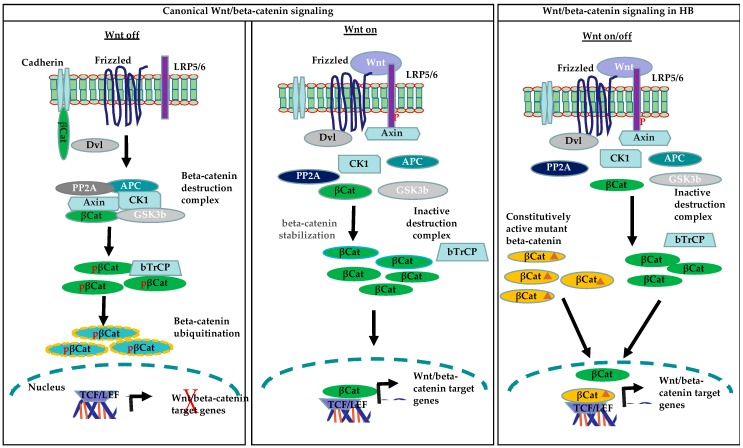
Accumulating evidence suggests that HB derives from less differentiated cells. Signals that promote self-renewal are one of the key events in the formation of cancer stem cells. Dysregulation of the developmental/self-renewal pathways in normal liver stem/progenitor cells plays a significant role in hepato-carcinogenesis. One of the most important developmental pathways associated with progenitor/stem cells is Wnt/beta-catenin, which is known to play an important role in the activation and expansion of progenitor/stem cells during embryogenesis and liver regeneration, and thus enables hepatic homeostasis [57,58]. Among human cancers tightly linked to abnormal Wnt/beta-catenin signaling, hepatoblastoma (HB) presents the highest rate (up to 90%) of beta-catenin mutations. The activation of the Wnt/beta-catenin signaling pathway is mediated by Wnt proteins, a family of secreted glycoproteins. Wnt ligands bind to cell membrane Frizzled (FZD) receptors and the co-receptor LRP 5/6 to mediate downstream activation of the Wnt/beta-catenin signaling pathway [59,60]. Wnt binding to Frizzled and LRP5/6 triggers the recruitment of the scaffolding protein Disheveled (Dvl), phosphorylation of LRP5/6 and Axin to the plasma membrane. [61]. The recruitment of Axin to the plasma membrane leads to the disruption of the beta-catenin destruction complex, which results in the stabilization and cytoplasmic accumulation of beta-catenin. Beta-catenin is then translocated to the nucleus, where it forms a complex with the T cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors to mediate the expression of Wnt target genes. In the absence of Wnt proteins, the level of beta-catenin is maintained at low levels through its degradation by the destruction complex. This complex consists of the proteins Axin, adenomatous polyposis coli (APC), glycogen synthase kinase 3 beta (GSK3beta), and casein kinase 1 alpha (CK1alpha). Axin, along with other components of the destruction complex, mediates the phosphorylation of beta-catenin at serine 45 by CK1alpha and phosphorylation of serine 33 and 37 and threonine 41 by GSK3beta. Under normal conditions, phosphorylated beta-catenin is recognized by E3 ubiquitin ligase beta-transducin repeat containing E3 ubiquitin protein ligase (beta-TrCP), which triggers the ubiquitination and proteasomal degradation of beta-catenin, thus maintaining its physiological levels in the cells [62]. Deletions of exon 3 of CTNNB1 or mutations prevent beta-catenin phosphorylation, and thus its degradation leads to an increase in both cytoplasmic and nuclear beta-catenin protein levels, which results in the constitutive activation of the Wnt/beta-catenin pathway (Figure 2).
Figure 2.
Canonical Wnt/beta-catenin and constitutively active beta-catenin signaling in HB (β-Cat: beta-catenin; pβ-Cat: phosphorylated beta-catenin; β-Cat: mutated beta-catenin)
Global analysis of HB tissues revealed a highly significant role of Wnt/beta-catenin in HB pathogenesis [14]. Oncoprotein Myc, one of the direct targets of the beta-catenin pathway, induced hepatoblastoma-like tumors in mice that resembled the human HB C2 subtype [13]. Genetic aberrations affecting exon 3 of CTNNB1 in the form of deletions or missense mutations that make beta-catenin stable and constitutively active were found to occur in HB at a rate of nearly 48% [19]. Additionally, Hepatocyte Growth Factor (HGF)/c-Met-mediated beta-catenin phosphorylation, which makes it highly active, was found in nearly 80% of HB tumors [63].
Recurrent mutations in CTNNB1 and increased expression of the transcription factor NFE2L2have been reported in HB [14]. Mutations in other components of the beta-catenin degradation complex, including those affecting APC and Axin have also been reported [19,64,65]. Beta-catenin-mediated expression of hepatic stem/progenitor markers and interplay of Wnt/beta-catenin and Myc signaling have been observed in aggressive HB [13]. A strong nuclear accumulation of both beta-catenin and Yes Associated Protein (YAP) has also been observed in HB tumor samples [66]. YAP is a transcription factor and an important component of the Hippo signaling pathway [67]. Constitutively active beta-catenin and YAP in hepatocytes led to the development of HB in experimental models, suggesting a functional crosstalk between beta-catenin and YAP in the pathogenesis of HB. Interestingly, a beta-catenin–YAP functional interaction was observed specifically in HB and not in HCC or intrahepatic cholangiocarcinoma samples [66]. YAP is activated in approximately 80% of human HB samples, as determined by its nuclear localization [66]. Valanejad et al. showed that the transcriptional regulator poly (ADP-ribose) polymerase 1 (PARP1) binds and activates aggressive liver cancer domains (ALCDs) in HB, which results in the induction of oncogenes, including beta-catenin [68]. Recently, Matsumoto et al. identified growth regulation by estrogen in breast cancer 1 (GREB1) as a novel downstream target of the Wnt/beta-catenin pathway that promoted HB cell growth, both in vitro and in vivo, by suppressing TGF beta signaling [69]. Even though other gene mutations are known to occur in HB, recent studies have demonstrated that genetic defects and aberrant activation of Wnt/beta-catenin signaling components play a significant role in the pathogenesis of HB. These defects can potentially lead to malignant transformation of normal hepatoblasts and eventually development of HB [19,64,70,71,72].
6. MicroRNA Regulation of Beta-Catenin Signaling in HB
MicroRNAs (miRNAs) are a family of small, endogenous, non-coding RNAs, approximately 19–25 nucleotides in length. MicroRNAs regulate key signaling pathways in the pathogenesis of hepatoblastoma and are known to regulate CSCs [73,74]. HB tissues have a high expression of miR-371-3 and decreased levels of miR-100/let-7a-2/miR-125b-1. Both of these miR clusters are differentially regulated by Myc, one of the direct targets of beta-catenin signaling [75]. MicroRNAs, such as let-7i-3p, miR-449b-3p, miR-624-5p, and miR-885-5p, downregulate Wnt signaling activity and inhibit HB cell growth. MicroRNAs miR-1246, miR-452, miR-217, miR-612, miR-214, miR-200a, and let7b have also been found to regulate Wnt signaling [76]. Let-7b suppresses the proliferation, invasion and migration of CSCs by inhibiting Wnt/beta-catenin signaling via downregulation of Frizzled 4 [77,78]. MicroRNA miR-1246 promotes the activation of Wnt/beta-catenin in CSCs by cooperating with Octamer 4, an upstream regulator of miR-1246 [79]. The miR-217-DKK1 axis promotes the stemness of cancer cells, resulting in the constitutive activation of Wnt signaling [80], whereas miR-612 suppresses the stemness of liver cancer cells through Wnt/beta-catenin signaling [81]. Ma et al. found that miR-130b facilitates the growth and self-renewal of CD133-positive liver tumor-initiating cells by targeting TP53INP1. Antagonizing miR-130b in CD133-positive tumor-initiating cells significantly increases sensitivity to chemotherapeutic agents and attenuates tumorigenicity in vivo [31]. The miR-200a is known to regulate epithelial–mesenchymal and stem-like transitions via zinc finger E-box binding homeobox 2 (ZEB2) and beta-catenin signaling, respectively [82].
7. Therapeutic Targeting of Cancer Stem Cells
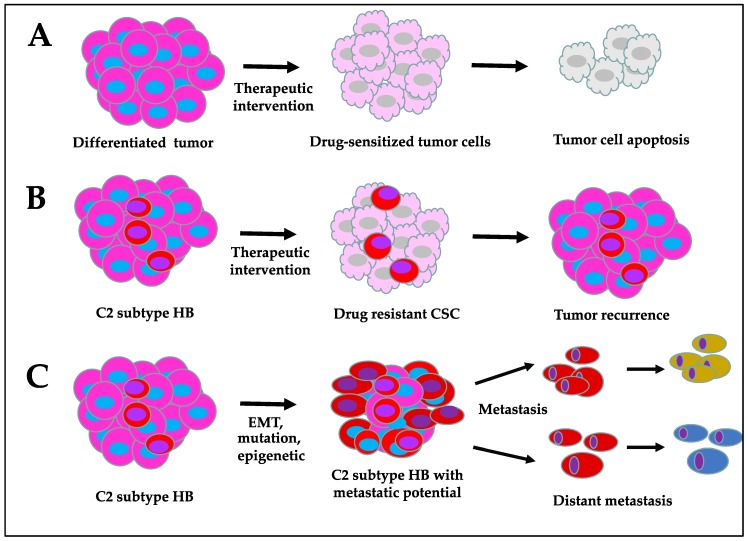
Targeting CSCs is challenging due to their high degree of heterogeneity and the intrinsic survival mechanisms that they develop over time to bypass apoptotic signals. At present, conventional therapies, such as chemotherapy, radiotherapy, and immunotherapy, can effectively suppress the growth of differentiated tumor cells; however, they potentially lag behind in inhibiting CSC proliferation (Figure 3). Although more advanced treatment options have become available in recent years, CSC resistance to therapies and cancer recurrence remain major challenges. CSCs develop therapeutic resistance via cellular reprogramming processes, such as EMT, altered drug tolerating/metabolizing ability, and DNA repair mechanisms, as well as their ability to resist apoptosis [83,84]. The accumulation of epigenetic/mutational changes and the influence by the tumor microenvironment signals may also contribute to chemoresistance. The surface markers of CSCs are generally shared by somatic progenitor cells. High expression of these markers has been observed in human HB tumors. The identification of differentially expressed CSC-specific surface markers, along with aberrantly expressed signaling targets and metabolic alterations, will hopefully aid in distinguishing CSCs from somatic progenitor cells, which may be exploited for selective CSC-targeted therapies. Targeting ATP-binding cassette (ABC) transporters, preventing CSC self-renewal and survival, transforming the more aggressive HB C2 subtype into C1 by inducing cellular differentiation, and developing CSC surface marker-targeting drug delivery approaches are some of the key areas of importance. To achieve these goals, it is necessary to have an in-depth knowledge of the biological characteristics of these cells with respect to their cellular origin, mechanism of propagation, and genetic and molecular defects that distinguish them from normal progenitor cells.
Figure 3.
Characteristics of cancer stem cell-enriched hepatoblastoma compared to differentiated tumor: (A) drug-sensitive differentiated tumor cells, (B) drug resistant CSC in HB, and (C) CSC-enriched metastatic HB.
Several basic and clinical studies have demonstrated the critical role for Wnt/beta-catenin signaling in HB. The greatest challenge in targeting Wnt/beta-catenin signaling is its complexity, as it consists of several components regulated by protein post-translational modifications at various levels, as well as extensive signaling crosstalk with other cell-signaling pathways. Targeting nuclear beta-catenin and/or its nuclear regulators responsible for the transcription of downstream Wnt/beta-catenin genes could be a plausible approach to target CSCs. Beta-catenin interaction with cyclic adenosine 3′,5′-monophosphate response element-binding protein (CBP) promotes stem cell renewal, while interaction with P300 enhances progenitor cell differentiation. Therefore, targeting the beta-catenin-CBP interaction is a promising strategy, and small-molecule drugs, such as ICG-001, have been found to effectively target cancer stem cells [39,85,86,87,88]. Inhibition of the beta-catenin–CBP interaction with ICG-001 targets CSCs but not normal cells, and has been reported to arrest cancer growth, both in vitro and in vivo [85,89]. A second-generation ICG-001 drug (PRI-724) is now in clinical trial (NCT01302405). Small-molecule drug FH535 prevents both Wnt and peroxisome proliferator-activated receptor (PPAR)-mediated signaling by suppressing the recruitment of beta-catenin co-activators to target gene promoters and has been shown to be active in different tumor cell lines [90]. PKF115-584, PKF118-310, and CGP049090 are inhibitors of the TCF/beta-catenin protein complex, binding to DNA target sequences [91]. They have been shown to induce apoptosis in vitro and suppress liver tumor growth in vivo regardless of the mutation status of CTNNB1 [92]. Activation of mammalian target of rapamycin complex 1 (mTORC1) has been reported in human HB cells and in a murine HB model driven by beta-catenin and YAP. Rapamycin, an inhibitor of mTORC1, has been found to significantly arrest HB growth in vivo by inhibiting mTORC1 activation, reducing proliferation, and altering the histology of HB from C2 to the more differentiated C1 subtype [93]. Based on these basic and clinical data, the Wnt/beta-catenin pathway and its interacting partners are some of the promising therapeutic targets that can be used to manage HB recurrence and metastasis.
8. Conclusions
Even though several cancer therapeutic approaches are available at present, tumor recurrence is a growing challenge in modern medicine. Recent evidences strongly suggest that CSCs that resist current therapies are the basis of cancer recurrence. Better understanding of the ways in which they originate from normal progenitor/stem or hepatoblast cells, their intrinsic self-renewal and survival mechanisms, and differential drug-metabolizing abilities compared to normal and differentiated cancer cells is critical for developing CSC-targeting therapies.
Funding
This work was supported by a seed grant (NM) from the Division of Applied Cell Biology and Physiology, Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Czauderna P., Lopez-Terrada D., Hiyama E., Häberle B., Malogolowkin M.H., Meyers R.L. Hepatoblastoma state of the art: Pathology, genetics, risk stratification, and chemotherapy. Curr. Opin. Pediatr. 2014;26:19–28. doi: 10.1097/MOP.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 2.Bell D., Ranganathan S., Tao J., Monga S.P. Novel advances in understanding of molecular pathogenesis of hepatoblastoma: A Wnt/Beta-catenin perspective. Gene Expr. 2017;17:141–154. doi: 10.3727/105221616X693639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linabery A.M., Ross J.A. Trends in childhood cancer incidence in the U.S. (1992–2004) Cancer. 2008;112:416–432. doi: 10.1002/cncr.23169. [DOI] [PubMed] [Google Scholar]
- 4.Tanimura M., Matsui I., Abe J., Ikeda H., Kobayashi N., Ohira M., Kaneko M., Yokoyama M. Increased risk of hepatoblastoma among immature children with a lower birth weight. Cancer Res. 1998;58:3032–3035. [PubMed] [Google Scholar]
- 5.Ikeda H., Matsuyama S., Tanimura M. Association between hepatoblastoma and very low birth weight: A trend or a chance? J. Pediatr. 1997;130:557–560. doi: 10.1016/S0022-3476(97)70239-7. [DOI] [PubMed] [Google Scholar]
- 6.McLaughlin C.C., Baptiste M.S., Schymura M.J., Nasca P.C., Zdeb M.S. Maternal and infant birth characteristics and hepatoblastoma. Am. J. Epidemiol. 2006;163:818–828. doi: 10.1093/aje/kwj104. [DOI] [PubMed] [Google Scholar]
- 7.Taat F., Bosman D.K., Aronson D.C. Hepatoblastoma in a girl with biliary atresia: Coincidence or co-incidence. Pediatr. Blood Cancer. 2004;43:603–605. doi: 10.1002/pbc.20115. [DOI] [PubMed] [Google Scholar]
- 8.Davies J.Q., De La Hall P.M., Kaschula R.O.C., Sinclair-Smith C.C., Hartley P., Rode H., Millar A.J.W. Hepatoblastoma—Evolution of management and outcome and significance of histology of the resected tumor. A 31-year experience with 40 cases. J. Pediatr. Surg. 2004;39:1321–1327. doi: 10.1016/j.jpedsurg.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Haas J.E., Muczynski K.A., Krailo M., Ablin A., Land V., Vietti T.J., Denman Hammond G. Histopathology and prognosis in childhood hepatoblastoma and hepatocarcinoma. Cancer. 1989;64:1082–1095. doi: 10.1002/1097-0142(19890901)64:5<1082::AID-CNCR2820640520>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 10.Czauderna P, Haeberle E., Hiyama A., Rangaswami M., Krailo R., Maibach E., Rinaldi Y., Feng D., Aronson M., Malogolowkin K., et al. The Children’s Hepatic Tumors International Collaboration (Chic): Novel global rare tumor database yields new prognostic factors in hepatoblastoma and becomes a research model. Eur. J. Cancer. 2016;52:92–101. doi: 10.1016/j.ejca.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo J.H., Ren B., Keryanov S., Tseng G.C., Rao U.N., Monga S.P., Strom S., Demetris A.J., Nalesnik M., Ranganathan S., et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valanejad L., Lewis K., Wright M., Jiang Y., D’Souza A., Karns R., Sheridan R., Gupta A., Bove K., et al. Fxr-Gankyrin axis is involved in development of pediatric liver cancer. Carcinogenesis. 2017;38:738–747. doi: 10.1093/carcin/bgx050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cairo S., Armengol C., De Reyniès A., Wei Y., Thomas E., Renard C.A., Pontoglio M., Pontoglio H., Strick-Marchand F., Levillayer Y., et al. Hepatic stem-like phenotype and interplay of Wnt/Beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Eichenmüller M., Trippel F., Kreuder M., Beck A., Schwarzmayr T., Häberle B., Kappler R., Cairo S., Leuschner I., Strom T.M., et al. The genomic landscape of hepatoblastoma and their progenies with Hcc-like features. J. Hepatol. 2014;61:1312–1320. doi: 10.1016/j.jhep.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Lee H., El Jabbour T., Ainechi S., Gay L.M., Elvin J.A., Vergilio J.A., Fabrizio D., Suh J., Ramkisson S.H., Ali S.M., et al. General paucity of genomic alteration and low tumor mutation burden in refractory and metastatic hepatoblastoma: Comprehensive genomic profiling study. Hum. Pathol. 2017;70:84–91. doi: 10.1016/j.humpath.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Curia M.C., Zuckermann M., De Lellis L., Catalano T., Lattanzio R., Aceto G., Mariani-Costantini R., Veschi S., Cama A., Otte J.-B., et al. Sporadic childhood hepatoblastomas show activation of beta-catenin, mismatch repair defects and p53 Mutations. Mod. Pathol. 2008;21:7–14. doi: 10.1038/modpathol.3800977. [DOI] [PubMed] [Google Scholar]
- 17.Udatsu Y., Kusafuka T., Kuroda S., Miao J., Okada A. High frequency of beta-catenin mutations in hepatoblastoma. Pediatr. Surg. Int. 2001;17:508–512. doi: 10.1007/s003830000576. [DOI] [PubMed] [Google Scholar]
- 18.Takayasu H., Horie H., Hiyama E., Matsunaga T., Hayashi Y., Watanabe Y., Ozaki T., Suita S., Kaneko M., Sasaki F., et al. Frequent deletions and mutations of the beta-catenin gene are associated with overexpression of cyclin D1 and fibronectin and poorly differentiated histology in childhood hepatoblastoma. Clin. Cancer Res. 2001;7:901–908. [PubMed] [Google Scholar]
- 19.Koch A., Denkhaus D., Albrecht S., Leuschner I., von Schweinitz D., Pietsch T. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Res. 1999;59:269–273. [PubMed] [Google Scholar]
- 20.Bläker H., Hofmann W.J., Rieker R.J., Penzel R., Graf M., Otto H.F. Beta-catenin accumulation and mutation of the CTNNB1 gene in hepatoblastoma. Genes Chromosomes Cancer. 1999;25:399–402. doi: 10.1002/(SICI)1098-2264(199908)25:4<399::AID-GCC14>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 21.Weber R.G., Pietsch T., von Schweinitz D., Lichter P. Characterization of genomic alterations in hepatoblastomas. A role for gains on chromosomes 8q and 20 as predictors of poor outcome. Am. J. Pathol. 2000;157:571–578. doi: 10.1016/S0002-9440(10)64567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park W.S., Oh R.R., Park J.Y., Kim P.J., Shin M.S., Lee J.H., Kim H.S., Lee S.H., Kim S.Y., et al. Nuclear localization of beta-catenin is an important prognostic factor in hepatoblastoma. J. Pathol. 2001;193:483–490. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH804>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 23.Crippa S., Ancey P.B., Vazquez J., Angelino P., Rougemont A.L., Guettier C., Meylan E., Zoete V., Delorenzi M., Michielin O. Mutant CTNNB1 and histological heterogeneity define metabolic subtypes of hepatoblastoma. EMBO Mol. Med. 2017;9:1589–1604. doi: 10.15252/emmm.201707814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comerford S.A., Hinnant E.A., Chen Y., Bansal H., Klapproth S., Rakheja D., Hammer R.E., Finegold M.J., Lopez-Terrada D., O’Donnell K.A., et al. Hepatoblastoma modeling in mice places Nrf2 within a cancer field established by mutant beta-catenin. JCI Insight. 2016;1:e88549. doi: 10.1172/jci.insight.88549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding Y., Xi Y., Chen T., Wang J.Y., Tao D.L., Wu Z.L., Li L., Li Y.-P., Li C., Zeng R. Caprin-2 enhances canonical Wnt signaling through regulating Lrp5/6 phosphorylation. J. Cell Biol. 2008;182:865–872. doi: 10.1083/jcb.200803147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia D., Dong R., Jing Y., Xu D., Wang Q., Chen L., Liu L., Li Q., Huang Y., Zhang Y., et al. Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology. 2014;60:1686–1696. doi: 10.1002/hep.27243. [DOI] [PubMed] [Google Scholar]
- 27.Cast A., Valanejad L., Wright M., Nguyen P., Gupta A., Zhu L., Timchenko N., Shin S. C/Ebpalpha-dependent preneoplastic tumor foci are the origin of hepatocellular carcinoma and aggressive pediatric liver cancer. Hepatology. 2018;67:1857–1871. doi: 10.1002/hep.29677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruck P., Xiao J.C. Stem-like cells in hepatoblastoma. Med. Pediatr. Oncol. 2002;39:504–507. doi: 10.1002/mpo.10175. [DOI] [PubMed] [Google Scholar]
- 29.Xiao Y., Lin M., Jiang X., Ye J., Guo T., Shi Y., Bian X. The recent advances on liver cancer stem cells: Biomarkers, separation, and therapy. Anal. Cell Pathol. 2017;2017:5108653. doi: 10.1155/2017/5108653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bahnassy A.A., Fawzy M., El-Wakil M., Zekri A.R.N., Abdel-Sayed A., Sheta M. Aberrant expression of cancer stem cell markers (Cd44, Cd90, and Cd133) contributes to disease progression and reduced survival in hepatoblastoma patients: 4-year survival data. Transl. Res. 2015;165:396–406. doi: 10.1016/j.trsl.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Ma S., Tang K.H., Chan Y.P., Lee T.K., Kwan P.S., Castilho A., Zheng B.J., Ng I., Man K., Zheng B.-J., et al. Mir-130b promotes Cd133+ liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694–707. doi: 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Ma S. Biology and clinical implications of Cd133+ liver cancer stem cells. Exp. Cell Res. 2013;319:126–132. doi: 10.1016/j.yexcr.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Rountree C.B., Senadheera S., Mato J.M., Crooks G.M., Lu S.C. Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A-deficient mice. Hepatology. 2008;47:1288–1297. doi: 10.1002/hep.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rountree C.B., Ding W., He L., Stiles B. Expansion of Cd133-expressing liver cancer stem cells in liver-specific phosphatase and tensin homolog deleted on chromosome 10-deleted mice. Stem Cells. 2009;27:290–299. doi: 10.1634/stemcells.2008-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mavila N., James D., Utley S., Cu N., Coblens O., Mak K., Wang K.S., Rountree B., Kahn M. Fibroblast growth factor receptor-mediated activation of akt-beta-catenin-cbp pathway regulates survival and proliferation of murine hepatoblasts and hepatic tumor initiating stem cells. PLoS ONE. 2012;7:e50401. doi: 10.1371/journal.pone.0050401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mavila N., James D., Shivakumar P., Nguyen M.V., Utley S., Mak K., Groff M., Wu A., Zhou S., Wang L., et al. Expansion of prominin-1-expressing cells in association with fibrosis of biliary atresia. Hepatology. 2014;60:941–953. doi: 10.1002/hep.27203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y.C., Yang J.Y., Yan L.N. Relevant markers of cancer stem cells indicate a poor prognosis in hepatocellular carcinoma patients: A meta-analysis. Eur. J. Gastroenterol. Hepatol. 2013;25:1007–1016. doi: 10.1097/MEG.0b013e32836019d8. [DOI] [PubMed] [Google Scholar]
- 38.Tang K.H., Ma S., Lee T.K., Chan Y.P., Kwan P.S., Tong C.M., Lo C.M., Ng I.O., Man K., To K.-F., et al. Cd133+ Liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/Cxcl1 signaling. Hepatology. 2012;55:807–820. doi: 10.1002/hep.24739. [DOI] [PubMed] [Google Scholar]
- 39.Tang Y., Berlind J., Mavila N. Inhibition of CREB binding protein-beta-catenin signaling down regulates Cd133 expression and activates PP2A-PTEN signaling in tumor initiating liver cancer cells. Cell Commun. Signal. 2018;16:9. doi: 10.1186/s12964-018-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma S., Lee T.K., Zheng B.J., Chan K.W., Guan X.Y. Cd133+ HCC Cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749–1758. doi: 10.1038/sj.onc.1210811. [DOI] [PubMed] [Google Scholar]
- 41.Stafman L.L., Williams A.P., Garner E.F., Aye J.M., Stewart J.E., Yoon K.J., Beierle E.A., Whelan K. Targeting Pim Kinases Affects Maintenance of Cd133 Tumor Cell Population in Hepatoblastoma. Transl. Oncol. 2019;12:200–208. doi: 10.1016/j.tranon.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita T., Budhu A., Forgues M., Wang X.W. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67:10831–10839. doi: 10.1158/0008-5472.CAN-07-0908. [DOI] [PubMed] [Google Scholar]
- 43.Yoon S.M., Gerasimidou D., Kuwahara R., Hytiroglou P., Yoo J.E., Park Y.N., Theise N.D. Epithelial cell adhesion molecule (EpCAM) marks hepatocytes newly derived from stem/progenitor cells in humans. Hepatology. 2011;53:964–973. doi: 10.1002/hep.24122. [DOI] [PubMed] [Google Scholar]
- 44.Dollé L., Theise N.D., Schmelzer E., Boulter L., Gires O., van Grunsven L.A. EpCAM and the biology of hepatic stem/progenitor cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;308:G233–G250. doi: 10.1152/ajpgi.00069.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka M., Okabe M., Suzuki K., Kamiya Y., Tsukahara Y., Saito S., Miyajima A. Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: Drastic change of EpCAM expression during liver development. Mech. Dev. 2009;126:665–676. doi: 10.1016/j.mod.2009.06.939. [DOI] [PubMed] [Google Scholar]
- 46.Maetzel D., Denzel S., Mack B., Canis M., Went P., Benk M., Gires O., Kieu C., Papior P., Baeuerle P.A., et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 2009;11:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 47.Yamashita T., Forgues M., Wang W., Kim J.W., Ye Q., Jia H., Tang Z.Y., Budhu A., Zanetti K.A., Chen Y., et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 48.Münz M., Murr A., Kvesic M., Rau D., Mangold S., Pflanz S., Rüttinger D., Lumsden J., Volkland J., Fagerberg J., et al. Side-by-side analysis of five clinically tested anti-EpCAM monoclonal antibodies. Cancer Cell Int. 2010;10:44. doi: 10.1186/1475-2867-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z.F., Ho D.W., Ng M.N., Lau C.K., Yu W.C., Ngai P., Fan S.T., Chu P.W., Lam C.T., Poon R.T.P. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 50.Singh A., Settleman J.E.M.T. Cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Campbell L.L., Brooks M., Reinhard F., Zhang C.C., et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ansieau S., Hinkal G., Thomas C., Bastid J., Puisieux A. Early origin of cancer metastases: Dissemination and evolution of premalignant cells. Cell Cycle. 2008;7:3659–3663. doi: 10.4161/cc.7.23.7049. [DOI] [PubMed] [Google Scholar]
- 53.Chen L., Tian X., Gong W., Sun B., Li G., Liu D., Song T., Guo P., He Y., Chen Z., et al. Periostin mediates epithelial-mesenchymal transition through the MAPK/ERK pathway in hepatoblastoma. Cancer Biol. Med. 2019;16:89–100. doi: 10.20892/j.issn.2095-3941.2018.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C., Liang Q.Y., Chen H.K., Wu P.F., Feng Z.Y., Ma X.M., Zhou G.Q., Wu H.R. Dram1 regulates the migration and invasion of hepatoblastoma cells via autophagy-EMT pathway. Oncol. Lett. 2018;16:2427–2433. doi: 10.3892/ol.2018.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z., Liu F., Yang F., Liu Y. Kockdown of Oip5-As1 Expression Inhibits Proliferation, Metastasis and Emt Progress in Hepatoblastoma Cells through up-Regulating Mir-186a-5p and Down-Regulating Zeb1. Biomed. Pharmacother. 2018;101:14–23. doi: 10.1016/j.biopha.2018.02.026. [DOI] [PubMed] [Google Scholar]
- 56.Fu X., Cui P., Chen F., Xu J., Gong L., Jiang L., Xiao Y., Zhang D. Thymosin beta 4 promotes hepatoblastoma metastasis via the induction of epithelial-mesenchymal transition. Mol. Med. Rep. 2015;12:127–132. doi: 10.3892/mmr.2015.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monga S.P. Beta-catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology. 2015;148:1294–1310. doi: 10.1053/j.gastro.2015.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Russell J.O., Monga S.P. Wnt/Beta-catenin signaling in liver development, homeostasis, and pathobiology. Annu. Rev. Pathol. 2018;13:351–378. doi: 10.1146/annurev-pathol-020117-044010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamai K., Zeng X., Liu C., Zhang X., Harada Y., Chang Z., He X.A. Mechanism for Wnt coreceptor activation. Mol. Cell. 2004;13:149–156. doi: 10.1016/S1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 60.MacDonald B.T., Tamai K., He X. Wnt/Beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamai K., Semenov M., Kato Y., Spokony R., Liu C., Katsuyama Y., He X., Hess F., Saint-Jeannet J.-P. Ldl-receptor-related proteins in wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 62.Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purcell R., Childs M., Maibach R., Miles C., Turner C., Zimmermann A., Sullivan M. Hgf/C-Met related activation of beta-catenin in hepatoblastoma. J. Exp. Clin. Cancer Res. 2011;30:96. doi: 10.1186/1756-9966-30-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koch A., Weber N., Waha A., Hartmann W., Denkhaus D., Behrens J., Pietsch T., Behrens J., Birchmeier W., von Schweinitz D. Mutations and elevated transcriptional activity of conductin (AXIN2) in hepatoblastomas. J. Pathol. 2004;204:546–554. doi: 10.1002/path.1662. [DOI] [PubMed] [Google Scholar]
- 65.Zhang L., Jin Y., Zheng K., Wang H., Yang S., Han W., Yang H., Yu Y., Yang Y., Geng D., et al. Whole-Genome Sequencing Identifies a Novel Variation of Was Gene Coordinating with Heterozygous Germline Mutation of Apc to Enhance Hepatoblastoma Oncogenesis. Front. Genet. 2018;9:668. doi: 10.3389/fgene.2018.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tao J., Calvisi D.F., Ranganathan S., Cigliano A., Zhou L., Singh S., Ribback S., Jiang L., Fan B., Terracciano L., et al. Activation of beta-catenin and yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;3:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao B., Li L., Lei Q., Guan K.L. The Hippo-YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valanejad L., Cast A., Wright M., Bissig K.D., Karns R., Weirauch M.T., Timchenko N. PARP1 activation increases expression of modified tumor suppressors and pathways underlying development of aggressive hepatoblastoma. Commun. Biol. 2018;1:67. doi: 10.1038/s42003-018-0077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsumoto S., Yamamichi T., Shinzawa K., Kasahara Y., Nojima S., Kodama T., Kikuchi A., Obika S., Takehara T., Morri E., et al. Greb1 induced by Wnt signaling promotes development of hepatoblastoma by suppressing TGFbeta signaling. Nat. Commun. 2019;10:3882. doi: 10.1038/s41467-019-11533-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aretz S., Koch A., Uhlhaas S., Friedl W., Propping P., Schweinitz D.V., Pietsch T. Should children at risk for familial adenomatous polyposis be screened for hepatoblastoma and children with apparently sporadic hepatoblastoma be screened for APC germline mutations? Pediatr. Blood Cancer. 2006;47:811–818. doi: 10.1002/pbc.20698. [DOI] [PubMed] [Google Scholar]
- 71.Mokkapati S., Niopek K., Huang L., Cunniff K.J., Ruteshouser E.C., DeCaestecker M., Huff V., Finegold M.J. Beta-catenin activation in a novel liver progenitor cell type is sufficient to cause hepatocellular carcinoma and hepatoblastoma. Cancer Res. 2014;74:4515–4525. doi: 10.1158/0008-5472.CAN-13-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang W., Yan H.X., Chen L., Liu Q., He Y.Q., Yu L.X., Chen C., Zhang S.H., Huang D.D., Tang L., et al. Wnt/Beta-catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells. Cancer Res. 2008;68:4287–4295. doi: 10.1158/0008-5472.CAN-07-6691. [DOI] [PubMed] [Google Scholar]
- 73.Cristóbal I., Sanz-Álvarez M., Luque M., Caramés C., Rojo F., García-Foncillas J. The Role of Micrornas in Hepatoblastoma Tumors. Cancers. 2019;11:409. doi: 10.3390/cancers11030409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lou W., Liu J., Gao Y., Zhong G., Ding B., Xu L., Fan W. MicroRNA regulation of liver cancer stem cells. Am. J. Cancer Res. 2018;8:1126–1141. [PMC free article] [PubMed] [Google Scholar]
- 75.Cairo S., Wang Y., de Reyniès A., Duroure K., Dahan J., Redon M.J., Buendia M.A., Fabre M., McClelland M., Wang X.W., et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc. Natl. Acad. Sci. USA. 2010;107:20471–20476. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Indersie E., Lesjean S., Hooks K.B., Sagliocco F., Ernault T., Cairo S., Grotzer M., Rullier A., Taque S., Guettier C., et al. MicroRNA therapy inhibits hepatoblastoma growth in vivo by targeting beta-catenin and Wnt signaling. Hepatol. Commun. 2017;1:168–183. doi: 10.1002/hep4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai H., Chen Y., Yang X., Ma S., Wang Q., Zhang Y., Yuan Y., Niu X., Ding G. Let7b Modulates the Wnt/Beta-catenin pathway in liver cancer cells via downregulated Frizzled4. Tumour Biol. 2017;39:1010428317716076. doi: 10.1177/1010428317716076. [DOI] [PubMed] [Google Scholar]
- 78.Jin B., Wang W., Meng X.X., Du G., Li J., Zhang S.Z., Fu Z.H., Zhou B.H. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer. 2016;16:863. doi: 10.1186/s12885-016-2904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chai S., Ng K.Y., Tong M., Lau E.Y., Lee T.K., Chan K.W., Wong N., Yuan Y.F., Cheung S.T., Wang X.Q., et al. Octamer 4/microRNA-1246 signaling axis drives Wnt/beta-catenin activation in liver cancer stem cells. Hepatology. 2016;64:2062–2076. doi: 10.1002/hep.28821. [DOI] [PubMed] [Google Scholar]
- 80.Jiang C., Yu M., Xie X., Huang G., Peng Y., Ren D., Kuang M., Lin M., Liu B., Liu M., et al. miR-217 targeting DKK1 promotes cancer stem cell properties via activation of the Wnt signaling pathway in hepatocellular carcinoma. Oncol. Rep. 2017;38:2351–2359. doi: 10.3892/or.2017.5924. [DOI] [PubMed] [Google Scholar]
- 81.Tang J., Tao Z.H., Wen D., Wan J.L., Liu D.L., Zhang S., Fan J., Cui J.F., Sun H.C., Wang L., et al. miR-612 suppresses the stemness of liver cancer via Wnt/Beta-catenin signaling. Biochem. Biophys. Res. Commun. 2014;447:210–215. doi: 10.1016/j.bbrc.2014.03.135. [DOI] [PubMed] [Google Scholar]
- 82.Xia H., Cheung W.K., Sze J., Lu G., Jiang S., Yao H., Lin M.C., Bian X.W., Poon W.S., Kung H. miR-200a regulates epithelial-mesenchymal to stem-like transition via ZEB2 and beta-catenin signaling. J. Biol. Chem. 2010;285:36995–37004. doi: 10.1074/jbc.M110.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Del Vecchio C.A., Feng Y., Sokol E.S., Tillman E.J., Sanduja S., Reinhardt F., Gupta P.B. De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling. PLoS Biol. 2014;12:e1001945. doi: 10.1371/journal.pbio.1001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gupta P.B., Pastushenko I., Skibinski A., Blanpain C., Kuperwasser C. Phenotypic plasticity: Driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell. 2019;24:65–78. doi: 10.1016/j.stem.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lenz H.J., Kahn M. Safely targeting cancer stem cells via selective catenin coactivator antagonism. Cancer Sci. 2014;105:1087–1092. doi: 10.1111/cas.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delgado E.R., Yang J., So J., Fanti M., Leimgruber S., Kahn M., Monga S.P., Ishitani T., Shin D., Wilson G.D. Identification and characterization of a novel small-molecule inhibitor of beta-catenin signaling. Am. J. Pathol. 2014;184:2111–2122. doi: 10.1016/j.ajpath.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McMillan M., Kahn M. Investigating Wnt signaling: A chemogenomic safari. Drug Discov. Today. 2005;10:1467–1474. doi: 10.1016/S1359-6446(05)03613-5. [DOI] [PubMed] [Google Scholar]
- 88.Emami K.H., Nguyen C., Ma H., Kim D.H., Jeong K.W., Eguchi M., Moon S.H., Moon R.T., Teo J.L., Kim H.Y., et al. A small molecule inhibitor of beta-catenin/creb-binding protein transcription [Corrected] Proc. Natl. Acad. Sci. USA. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eguchi M., Nguyen C., Lee S.C., Kahn M. Icg-001, a Novel Small Molecule Regulator of Tcf/Beta-Catenin Transcription. Med. Chem. 2005;1:467–472. doi: 10.2174/1573406054864098. [DOI] [PubMed] [Google Scholar]
- 90.Handeli S., Simon J.A. A small-molecule inhibitor of Tcf/beta-catenin signaling down-regulates Ppargamma and Ppardelta Activities. Mol. Cancer Ther. 2008;7:521–529. doi: 10.1158/1535-7163.MCT-07-2063. [DOI] [PubMed] [Google Scholar]
- 91.Lepourcelet M., Chen Y.N.P., France D.S., Wang H., Crews P., Petersen F., Shivdasani R.A., Bruseo C., Wood A.W. Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell. 2004;5:91–102. doi: 10.1016/S1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 92.Wei W., Chua M.S., Grepper S., So S. Small molecule antagonists of Tcf4/Beta-catenin complex inhibit the growth of HCC cells in vitro and in vivo. Int. J. Cancer. 2010;126:2426–2436. doi: 10.1002/ijc.24810. [DOI] [PubMed] [Google Scholar]
- 93.Molina L., Yang H., Michael A.O.A., Oertel M., Bell A., Singh S., Monga S.P., Chen X., Tao J. mTOR inhibition affects Yap1-Beta-catenin-induced hepatoblastoma growth and development. Oncotarget. 2019;10:1475–1490. doi: 10.18632/oncotarget.26668. [DOI] [PMC free article] [PubMed] [Google Scholar]