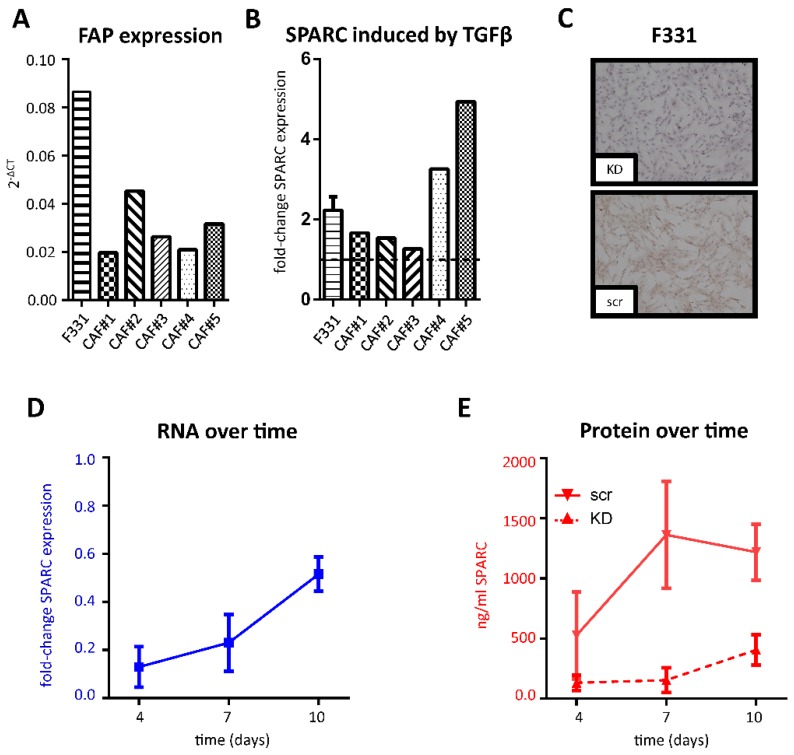
Figure 2.
Fibroblast-derived SPARC: RNA was isolated from semiconfluent fibroblast cultures and FAP mRNA was analyzed by qRT-PCR relative to GAPDH as the housekeeping gene (A). F331 and CAFs were treated with 5 ng/mL TGFβ for a total of 72 h. SPARC mRNA expression was plotted as fold change to negative control and shown for F331 fibroblasts (n = 3) and for CAFs (single experiments); numbering of CAFs indicate the respective patient from whom the fibroblasts were isolated (B). In addition, F331 cells transfected with SPARC siRNA or scr control were grown on glass slides and stained by IHC with a SPARC specific mAB after 72 h (C); size bar = 100 µm. Knockdown efficiency in F331 cells was monitored over a total of 10 days (D) SPARC mRNA expression after siRNA transfection was calculated as fold-change ratio of KD to scr (n = 3). (E) SPARC protein levels secreted into the medium supernatant were evaluated by ELISA and plotted for both scr (solid line) and KD (dashed line) (n = 2). All data are pooled from experiments using 2 different siRNAs and given as mean ± SD. KD, knockdown; scr, scrambled control; siRNA, small interfering RNA.