Abstract
Reciprocal epithelial–mesenchymal interactions are pivotal in lung development, homeostasis, injury, and repair. Organoids have been used to investigate such interactions, but with a major focus on epithelial responses to mesenchyme and less attention to epithelial effects on mesenchyme. In the present study, we used nascent organoids composed of human and mouse lung epithelial and mesenchymal cells to demonstrate that healthy lung epithelium dramatically represses transcriptional, contractile, and matrix synthetic functions of lung fibroblasts. Repression of fibroblast activation requires signaling via the bone morphogenetic protein (BMP) pathway. BMP signaling is diminished after epithelial injury in vitro and in vivo, and exogenous BMP4 restores fibroblast repression in injured organoids. In contrast, inhibition of BMP signaling in healthy organoids is sufficient to derepress fibroblast matrix synthetic function. Our results reveal potent repression of fibroblast activation by healthy lung epithelium and a novel mechanism by which epithelial loss or injury is intrinsically coupled to mesenchymal activation via loss of repressive BMP signaling.
Keywords: fibrosis, mesenchymal, coculture, airway, alveolar
Epithelial–mesenchymal interactions are pivotal during tissue development and organ morphogenesis (1). These interactions continue in mature organisms, helping to maintain tissue homeostasis, self-renewal, and healing response to injury (2). In the lung, epithelial–mesenchymal interactions are essential to the formation of the branching airways and alveolarization of the distal lung (3, 4). Epithelial injury to both the proximal airways and the distal alveoli generates mesenchymal cell activation as part of the normal wound-healing response (5–7). In most cases, this process is self-limiting, leading to tissue repair and injury resolution. However, in both the proximal airways and distal alveoli, persistent epithelial injury or aberrant repair leads to continued mesenchymal activation, scarring, and loss of lung function (5–12). Thus, understanding the specific signals that mediate epithelial–mesenchymal cross-talk during tissue homeostasis, normal tissue repair, and fibrotic tissue remodeling is a high priority. Accumulating evidence, largely generated using mouse models, has linked epithelial–mesenchymal interactions coordinated by multiple pathways, including hedgehog, fibroblast growth factor, and Wnt signaling, to lung homeostasis and fibrosis (3, 13–15). Although these efforts represent important advances, it remains technically challenging and time consuming to precisely manipulate and dissect epithelial–mesenchymal interactions in mouse model systems, motivating efforts to study these interactions in additional physiologically relevant model systems.
Traditional cell culture systems, including Transwell systems, have also been used to study epithelial–mesenchymal interactions, and they have similarly shown that epithelial cell injury activates fibroblasts (12, 16–19). However, such investigations have focused overwhelmingly on injury responses, with little attention focused on homeostatic interactions between epithelial cells and fibroblasts. Organoids have recently emerged as valuable new tools to understand cell–cell interactions and cell differentiation and as models for studying disease processes. Organoids form and function stably over a period of days and weeks through a process of self-organization, offering an opportunity to study cell–cell interactions critical to determining cell fate and homeostasis. In particular, investigations of organoids formed from lung cells or from pluripotent progenitors have probed the mechanisms controlling epithelial lineage commitment and differentiation into mature cell types (20–28), and they have highlighted the role that the mesenchyme can play in supporting such processes. In comparison, less attention has been directed at studying the influence of the epithelium on mesenchymal cell states within such organoid systems. Given the key role that lung mesenchyme activation plays in wound healing and fibrosis, in this work we set out to study how mesenchymal activation is influenced by lung epithelium within nascent organoids and how it is altered in response to epithelium- and fibroblast-specific perturbations. Using our previously developed airway organoid model (29), we focused on early epithelial–mesenchymal interactions during nascent organoid emergence to demonstrate potent epithelial repression of fibroblast transcriptional, contractile, and matrix synthetic states, in part through bone morphogenetic protein (BMP) signaling. We found that epithelial injury alone is sufficient to recapitulate mesenchymal activation, whereas healthy epithelium retains the capacity to repress exogenous fibroblast-specific activation. Together these results demonstrate that healthy epithelial–mesenchymal cross-talk normally acts to restrain fibroblast contractile and matrix synthetic activation, and they establish this system as a powerful tool for dissecting the signals that underlie homeostatic, reparative, or fibrotic interactions between epithelial and mesenchymal compartments.
Methods
Full details of the methods are provided in the data supplement.
Mice and Bleomycin-induced Lung Injury
All experiments were performed in accordance with the guidelines of the Mayo Clinic Institutional Animal Care and Use Committee. Col1a1 (collagen type I alpha 1 chain)-GFP transgenic mice were kindly provided by Dr. Derek Radisky and generated as previously described (30). Bleomycin (APP Pharmaceutical) was delivered to the lungs as previously described (31).
Cell Sorting and Culture
Mouse lung fibroblasts were isolated from Col1a1-GFP+ mice. Mouse type 2 alveolar epithelial cells were isolated from Sftpc-EGFP+ (surfactant protein C–enhanced GFP–positive) mice [B6N.Cg-Tg(Sftpc,-EGFP)1Dobb/J, CBG], which were purchased from The Jackson Laboratory (32).
C57BL/6 mouse primary tracheal and bronchial epithelial cells (mTEs; Cell Biologics) were cultured in Complete Epithelial Cell Medium (Cell Biologics). Normal human lung fibroblasts (hLFs; Lonza) were cultured in Dulbecco’s modified Eagle medium supplemented with 10% FBS and 1% antibiotic-antimycotic solution. Normal bronchial epithelial cells (Lonza) were cultured in BEGM BulletKit bronchial epithelial cell growth medium (Lonza). For two-dimensional (2D) coculture experiments, 3 × 105 mTEs and 3 × 105 hLFs were seeded into a well of a 6-well plate. Epithelial cell medium was used to maintain both cell types. Organoids were generated as described in prior work (29).
RNA Extraction and RT-PCR Analysis
Cells in the three-dimensional (3D) organoid were removed from Matrigel with Corning Cell Recovery Solution (Corning). Total RNA was isolated with the RNeasy Plus Mini Kit (Qiagen) or the RNeasy Micro Kit (Qiagen), depending on the number of cells. cDNA was synthesized and gene expression levels quantified by qRT-PCR using species-specific primers for all genes.
RNA-Sequencing Analysis of the Sorted Cell Population
RNA libraries were prepared and sequenced following Illumina’s standard protocol using the Illumina cBot and HiSeq 3000/4000 PE Cluster Kit, yielding 33 million–40 million fragment reads per sample. Differentially expressed genes of sorted Col1a1-GFP+ fibroblasts in the presence or absence of epithelial cells were identified using Smyth’s moderated t test and the Benjamini-Hochberg procedure for adjusted P values (false discovery rate). Genes with a false discovery rate below 0.05, absolute log2 fold change greater than 1, and annotated by Ensembl as being protein-coding genes were defined as being differentially expressed.
Immunofluorescence Microscopy
Mouse lungs were inflated and fixed with 4% paraformaldehyde (Polysciences). Organoids were removed and separated from the Matrigel layer with Corning Cell Recovery Solution, embedded in optical cutting temperature compound, and frozen at −80°C. Cryosections (10 μm) were placed on poly-l-lysine–coated slides (Fisher Scientific) for immunostaining with primary antibodies for FITC-conjugated ACTA2 (Sigma-Aldrich) diluted 1:200 or for BMP4 antibody (Santa Cruz Biotechnology) diluted 1:50 at 4°C overnight.
Western Blotting
Cells in 3D organoids were removed from Matrigel with Corning Cell Recovery Solution and were harvested into radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific) with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Lysates were subjected to SDS-PAGE, then transferred and probed with GAPDH (Cell Signaling Technology), ACTA2 (Sigma-Aldrich), and fibronectin (Santa Cruz Biotechnology) antibodies.
BMP Luciferase Assay
Cells were transfected with pGL3 BRE Luciferase (plasmid 45126; Addgene) (33) using Lipofectamine 3000 reagent (Life Technologies). Luciferase activity was measured via a dual luciferase reporter assay (Promega), with firefly luciferase activity normalized to Renilla activity.
Statistical Analysis
All results are expressed as mean ± SEM. Comparisons between groups were made by using an unpaired two-tailed Student’s t test. The statistics were analyzed using Prism 7 software (GraphPad Software). P values less than 0.05 were considered statistically significant. All experiments were repeated at least three times, and representative data are shown.
Results
Organoids Formed from Either Proximal or Distal Lung Epithelium Strongly Repress Fibroblast Activation Gene Signature
To evaluate the capacity for epithelial cells to regulate fibroblasts in vitro, we adapted a previously described organoid system (29). We previously showed that in this system, primary lung epithelial cells and fibroblasts rapidly condense, self-organize, and undergo stable epithelial maturation into both proximal and distal fates while seeded on a bed of Matrigel and cultured in the presence of lung epithelial differentiation factors (29). To enable efficient study of epithelial–mesenchymal interactions and responses to prior epithelium- or fibroblast-specific perturbations, we focused on early time points (Days 3–7) after cell seeding, and we term these cultures “nascent organoids,” reflecting their relatively early stage of self-organization and maturation (29).
To unambiguously distinguish fibroblast from epithelial transcriptional responses, we combined C57BL/6 mTEs and hLFs in these cultures, and we designed primers that amplified fibroblast and housekeeping gene expression in a highly species-specific manner (see Table E1 in the data supplement). For comparison, we cultured fibroblasts alone under identical conditions atop Matrigel and in the presence of the same epithelial differentiation factors (Figure 1A). Under these conditions, fibroblasts also rapidly condense into a single mass. To ascertain whether the epithelial regulation of fibroblasts was unique to the culture conditions, we also cultured hLFs alone or in the presence of mTEs in the same ratios as in organoids, but on traditional tissue culture plastic.
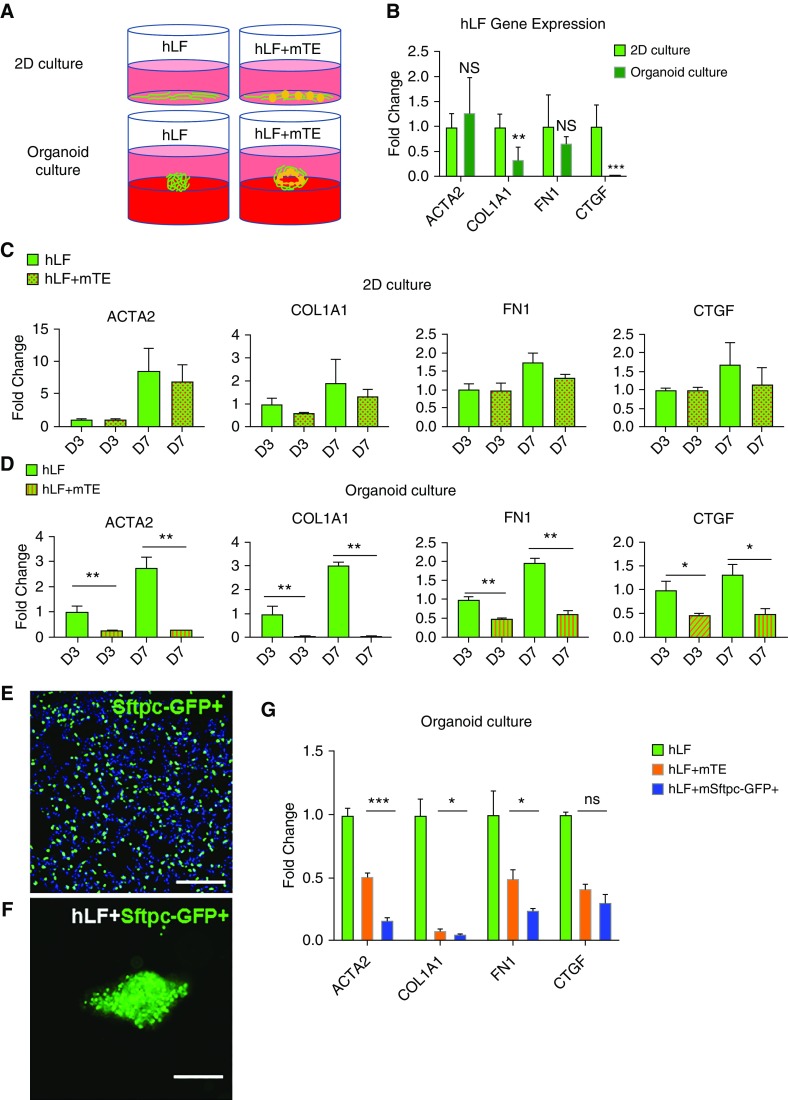
Figure 1.
Organoid culture with proximal or distal lung epithelium represses expression of a fibroblast activation gene signature. (A) Schematic diagram of two-dimensional (2D) and organoid cultures of fibroblasts alone or with epithelial cell–fibroblast cocultures. (B) Gene expression analysis of markers of fibroblast activation in 2D and organoid culture conditions for 3 days. n = 6. (C) Gene expression analysis of fibroblast transcripts in 2D mono- and cocultures at Day 3 and Day 7. n = 6. (D) Gene expression analysis of fibroblast transcripts in organoid mono- and cocultures at Day 3 and Day 7. n = 6. (E) Visualization of Sftpc-GFP+ cells within a transgenic mouse lung section. Scale bar: 100 μm. (F) Visualization of Sftpc-GFP+ cells within an epithelial cell–fibroblast organoid coculture. Scale bar: 100 μm. (G) Gene expression analysis of fibroblast transcripts in organoid culture conditions from fibroblasts alone versus in cocultures with mouse primary tracheal and bronchial epithelial cells (mTEs) or Sftpc-GFP+ alveolar epithelial cells at Day 3. n = 3. *P < 0.05, **P < 0.01, and ***P < 0.001. COL1A1 = collagen type I α 1 chain; CTGF = connective tissue growth factor; FN1 = fibronectin 1; hLFs = normal human lung fibroblasts; NS = not significant; Sftpc = surfactant protein C.
To first assess the baseline state of the fibroblast-only cultures, we compared their expression of a panel of genes characteristic of fibroblast activation (8, 14, 34). Fibroblasts alone cultured in the traditional 2D tissue culture plastic system exhibited higher expression of COL1A1 and CTGF (connective tissue growth factor), but not of ACTA2 or FN1 (fibronectin 1), than fibroblasts seeded atop 3D Matrigel (Figure 1B), demonstrating the relatively modest effect of culture system alone on fibroblasts. Inclusion of epithelial cells in coculture with fibroblasts in 2D did not significantly alter fibroblast expression of this panel of genes at either 3 or 7 days (Figure 1C). In contrast, in the organoid coculture model, the presence of epithelial cells significantly lowered expression of all four fibroblast activation genes across both Days 3 and 7 compared with cultures containing hLFs alone (Figure 1D). This response was particularly impressive for COL1A1 (0.063 ± 0.005-fold; P < 0.01) and ACTA2 (0.255 ± 0.014-fold; P < 0.01). To extend these observations of proximal epithelial suppression of fibroblast state to distal alveolar epithelial cells, we isolated type 2 alveolar epithelial cells from adult Sftpc-GFP mice by FACS (Figure 1E) and cocultured these cells with hLFs under identical seeding and culture conditions (Figure 1F). Fibroblasts grown in 3D organoids with Sftpc-GFP+ cells exhibited similar, and in three of four cases statistically greater, reductions in expression of the fibrotic activation gene panel (Figure 1G). These results highlight the dramatic influence that both proximal and distal lung epithelial cells exert on fibroblast gene expression, particularly when cultured under organoid-forming conditions, and they suggested that the fibrogenic and contractile state of fibroblasts is strongly influenced by proximity to, and reciprocal interactions with, lung epithelium.
Organoid Culture Restrains Fibroblast Proliferation, Contraction, and Extracellular Matrix Deposition
Functional readouts of fibroblast activation include proliferation, contractile capacity, and extracellular matrix (ECM) deposition. Previous studies have shown that organoid self-organization, compaction, and formation atop Matrigel require mechanical forces, particularly those generated by contractile fibroblasts (35, 36). In general agreement with epithelial cells restraining fibroblast contractile capacity, the compacted size of epithelial fibroblast organoids was significantly larger in diameter than that of fibroblasts alone, inclusive of whether fibroblast seeding density was maintained or doubled to be equivalent to the total cell density in epithelial cell–fibroblast cocultures (Figures 2A–2D). These results are consistent with epithelial cells reducing fibroblast contractile function and therefore limiting the fibroblast-mediated compaction needed to form dense cell aggregates during organoid formation. A potential alternative explanation is that cells in coculture proliferate more rapidly than in fibroblast-only cultures, leading to larger cell aggregates. However, when we visualized protein expression of the proliferative cell marker Ki-67 by immunofluorescence staining, we observed fewer Ki-67+ cells in epithelial–mesenchymal cocultures (Figure 2F) than we did in fibroblast-only cultures (Figure 2E), with a quantitative reduction in Ki-67+/vimentin+ fibroblasts in cocultures (Figure 2G). These results demonstrate that epithelial cells repress fibroblast proliferation in nascent organoids and rule out the possibility that the larger cell aggregate size is generated by greater proliferation.
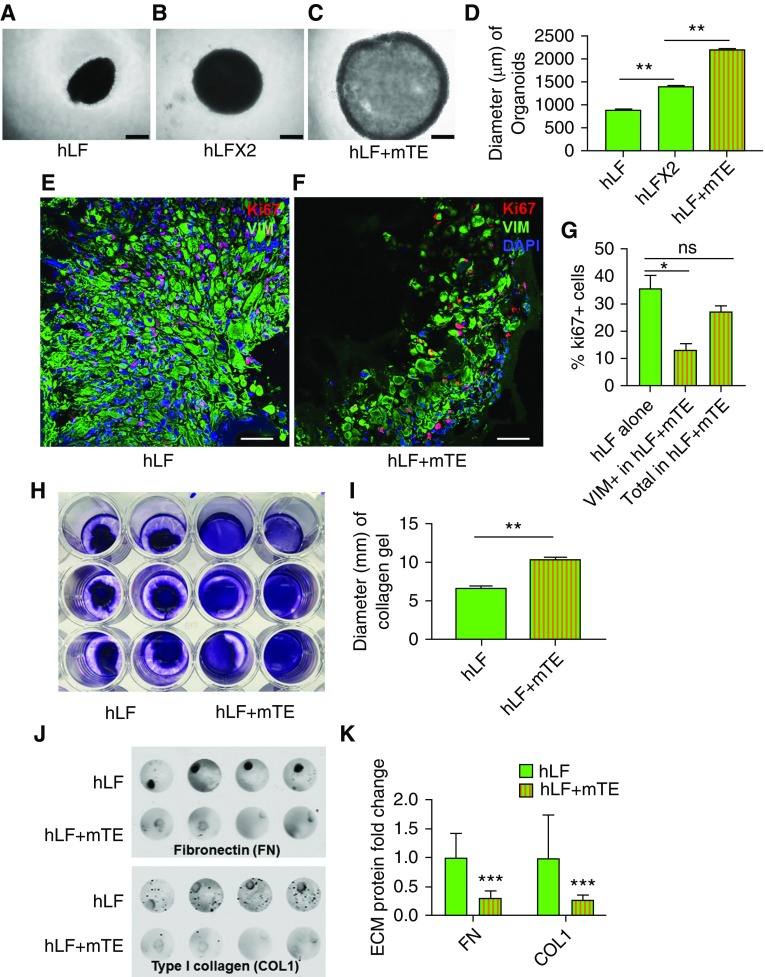
Figure 2.
Organoid culture restrains fibroblast proliferation, contractile function, and extracellular matrix (ECM) deposition. (A) Bright-field microscopy of fibroblasts (1 × 105 hLF) alone in organoid culture conditions. Scale bar: 500 μm. (B) Bright-field microscopy of fibroblasts at twice the seeding density of A (2 × 105 hLF) in organoid culture conditions. Scale bar: 500 μm. (C) Bright-field microscopy of coculture organoids (1 × 105 hLF and 1 × 105 mTEs), with total cell number the same as in B. Scale bar: 500 μm. (D) Quantification of three-dimensional organoid sizes. n = 5. (E–G) Immunofluorescence imaging of Ki-67 staining in human lung fibroblasts (vimentin-positive [VIM+] cells) from organoid-cultured fibroblasts alone (hLF; E) and cocultures (hLF + mTE; F). Scale bar: 50 μm. (G) Quantification of Ki-67+ cells in organoid-cultured fibroblasts alone and cocultures (Vim+ fibroblasts and total cells). n = 3. (H) Collagen gel contraction assay with cells labeled using trypan blue staining. (I) Quantification of collagen gel size showing greater gel compaction by fibroblasts alone and reduced compaction in the presence of mTEs. n = 6. (**P < 0.01). (J) Immunodetection of deposited fibronectin and collagen I in organoid cultures of fibroblasts alone (hLF) and cocultures (hLF + mTE). (K) Quantification of ECM deposition of fibronectin and collagen I. n = 8. *P < 0.05, **P < 0.01, and ***P < 0.001.
To test the effect of epithelial cells on fibroblast contractility directly, we suspended fibroblasts alone or in coculture with epithelial cells within 3D collagen hydrogels and studied the overall gel compaction over 24 hours (Figure 2H). These studies confirmed that mTEs significantly impaired collagen gel compaction (Figure 2I), again consistent with epithelial cells exerting a restraining effect on fibroblast contractile capacity. These findings are in agreement with prior work using human bronchial epithelial cells in a similar fibroblast collagen compaction system (37).
To evaluate the capacity of epithelial cells to reduce the matrix synthetic function of organoid cocultured fibroblasts, we used an immunofluorescence assay to quantify the deposition of ECM proteins fibronectin and collagen type I. Compared with fibroblasts alone, organoid cocultures exhibited significantly lower deposition of both ECM proteins (Figures 2J and 2K). Taken together, these results confirm significant functional effects of lung epithelium in repressing fibroblast proliferative, contractile, and fibrogenic phenotypes.
Organoid Culture Dampens Fibroblast Activation by Transforming Growth Factor-β1
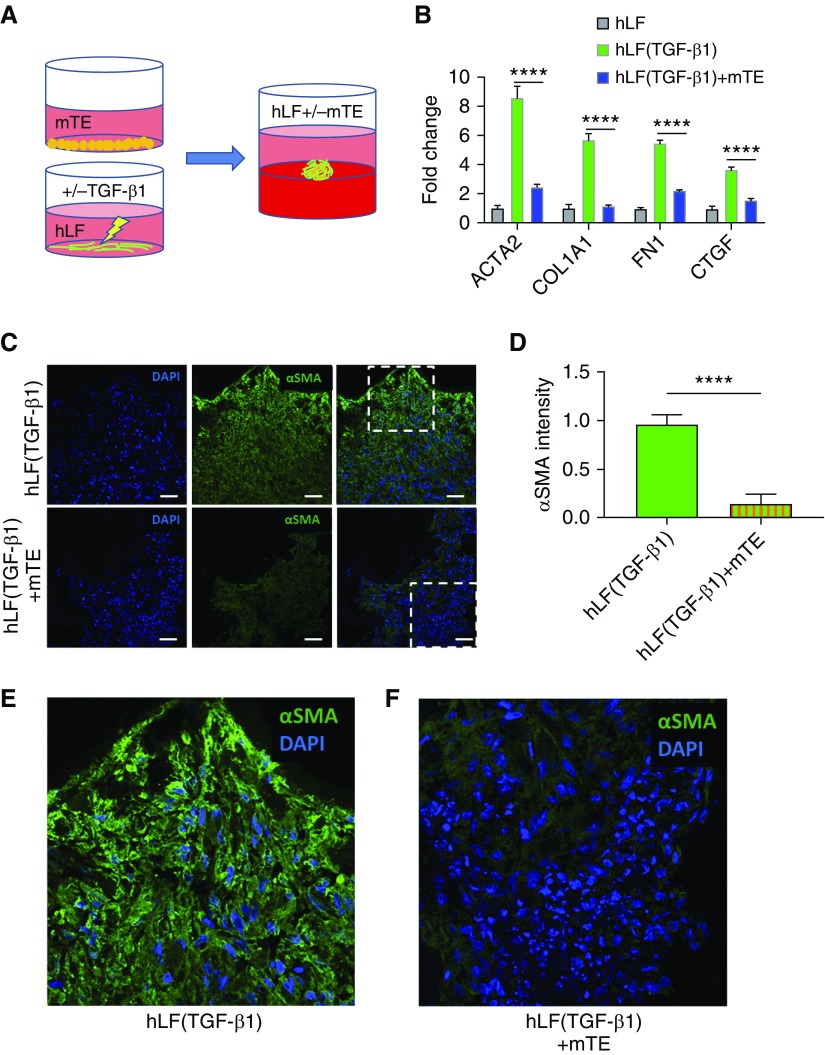
Although the experiments described above demonstrated that epithelial cells in organoid cocultures could repress fibroblast genetic and functional activation signatures, these fibroblasts were otherwise not exogenously activated. In prior work, we observed that transforming growth factor (TGF)-β1, the prototypical profibrotic cytokine known to activate fibroblasts to a contractile myofibroblast and matrix synthetic state, exerts profibrogenic effects even in the presence of epithelium (29). However, TGF-β1 in the lung can exert its profibrotic effects through either epithelium- or fibroblast-specific signaling (38, 39). To specifically test whether unperturbed epithelial cells (not exposed to TGF-β1) maintained a capacity to restrain fibroblast activation, we pretreated fibroblasts with TGF-β1 (5 ng/ml) for 24 hours in monolayer culture, then used these cells alone or mixed with mTEs cells to form organoids atop Matrigel for 72 hours as previously described (Figure 3A). Despite the preactivated state of the TGF-β1–stimulated fibroblasts, epithelial cells maintained a significant capacity to repress the fibroblast activation gene signature (Figure 3B). As a further readout of TGF-β1–mediated fibroblast activation, we visualized ACTA2, the prototype myofibroblast cytoskeletal protein, by immunofluorescence staining. Again, fibroblasts alone pretreated with TGF-β1 exhibited abundant ACTA2 staining that was significantly reduced when TGF-β1–pretreated fibroblasts were cocultured in organoids with mTEs (Figures 3C–3F and E1). These results demonstrate that healthy unperturbed epithelial cells retain the capacity to repress fibroblast activation even when fibroblasts are pretreated to an activated state by exposure to TGF-β1, further emphasizing the capacity of the healthy epithelium to restrain fibrogenic fibroblast activation.
Figure 3.
Organoid culture represses fibroblasts preactivated with transforming growth factor (TGF)-β1. (A) Schematic showing pretreatment of normal hLF alone with TGF-β1 followed by organoid culture of these cells alone (with or without TGF-β1 pretreatment) or cocultured with mouse primary mTEs. (B) Gene expression analysis of fibroblast transcripts in organoid culture conditions after fibroblast-only pretreatment with or without TGF-β1 and mono- or coculture with mTEs for 3 days. n = 6. (C) Immunofluorescence imaging of ACTA2 (αSMA) staining in fibroblasts pretreated with TGF-β1 followed by organoid culture as hLF alone (top) or in hLF + mTE cocultures. Scale bars: 50 μm. (D) Quantification of ACTA2 intensity in fibroblasts pretreated with TGF-β1 and transferred to organoid culture of fibroblasts alone or in coculture with mTEs. n = 6. (E) Enlarged detail from top row of C. (F) Enlarged detail from bottom row of C. ****P < 0.0001. α-SMA = α-smooth muscle actin.
Epithelial Injury Impairs Fibroblast Repression
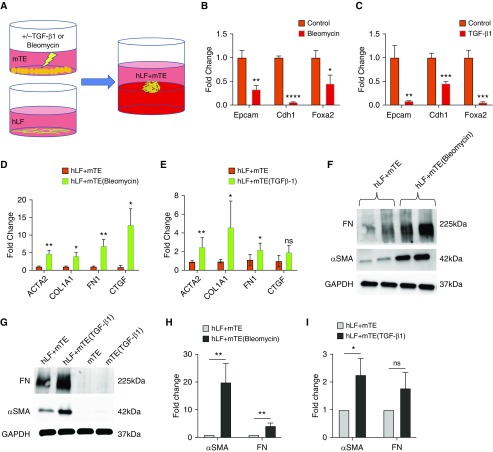
Because epithelial injury is often associated with the initiation and progression of fibrotic tissue remodeling (5–7, 9, 10), we next addressed the converse situation of an epithelium preinjured and then cocultured in organoids with unperturbed fibroblasts. We pretreated mTEs with bleomycin (15 μg/ml), a chemotherapeutic agent widely used to initiate lung epithelial injury in experimental models of pulmonary fibrosis (40), or by stimulation with TGF-β1 (5 ng/ml) (Figure 4A), a cytokine capable of initiating lung fibrogenesis exclusively through epithelial signaling (39). We confirmed that epithelial pretreatment with either bleomycin or TGF-β1 for 24 hours perturbed the epithelial state, as measured by reduced expression of epithelial differentiation markers Epcam (epithelial cellular adhesion molecule), Cdh1 (E-cadherin), and Foxa2 (Forkhead box A2) (Figures 4B and 4C) compared with untreated epithelial cells. Pretreatment of epithelial cells with bleomycin before organoid coculture with fibroblasts significantly elevated fibroblast expression of ACTA2, COL1A1, FN1, and CTGF (Figure 4D) at 48 hours. Similarly, pretreatment of epithelial cells only with TGF-β1 increased fibroblast expression of ACTA2, COL1A1, and FN1 significantly in organoid coculture (Figure 4E). Western blotting confirmed increased profibrotic protein expression of ACTA2 and fibronectin in organoids pretreated with bleomycin (Figures 4F and 4H) and TGF-β1 (Figures 4G and 4I) before organoid culture. In contrast, mTEs cultured alone with or without TGF-β1 prestimulation did not generate measurable amounts of either ACTA2 or fibronectin protein expression (Figure 4G), indicating that the major source of these proteins in the organoid system is fibroblasts. Taken together, these results indicate that healthy epithelial cells potently repress fibroblast activation and that epithelial injury alone is sufficient to provoke fibroblast activation. Although prior researchers have long sought to identify profibrotic factors released by lung epithelium in the setting of injury (6, 9–12, 41), the cumulative evidence presented in this article strongly supports an additional or alternative concept that active signaling processes repress fibroblast contractile and matrix synthetic states and maintain epithelial–mesenchymal homeostasis in the absence of injury.
Figure 4.
Injured epithelial cells no longer repress fibroblast activation. (A) Schematic diagram of mTEs pretreated with bleomycin or TGF-β1 and then cocultured with hLF. (B) Gene expression analysis of lung epithelial cells after exposure to bleomycin and before organoid coculture. n = 6. (C) Gene expression analysis of lung epithelial cells after exposure to TGF-β1 and before organoid coculture. n = 6. (D) Gene expression analysis of fibroblast transcripts in organoid cocultures formed with control and bleomycin-pretreated mTEs. n = 6. (E) Gene expression analysis of fibroblast transcripts in organoid cocultures formed with control and TGF-β1–pretreated mTEs. n = 5. (F) Western blots showing elevated FN and ACTA2 (αSMA) protein expression in organoids formed with epithelial cells preinjured with bleomycin. (G) Western blots showing elevated FN and ACTA2 protein expression in organoids formed from epithelial cells pretreated with TGF-β1 and lack of staining from mTEs alone. (H and I) Quantification of Western blots showing increased FN and ACTA2 protein expression in organoids formed with mTEs preinjured with bleomycin (H; n = 4) and TGF-β1 (I; n = 3) compared with organoids formed with control mTEs. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
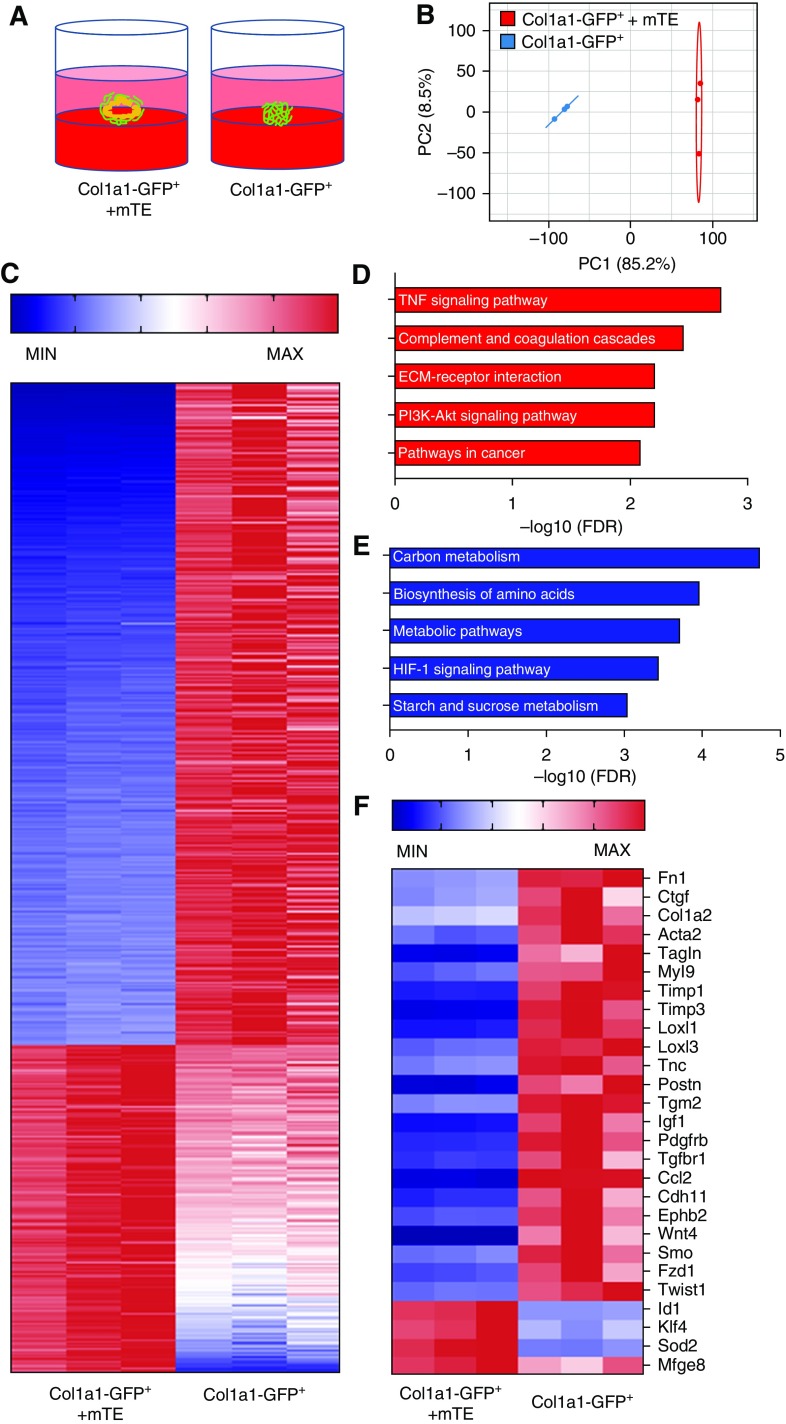
RNA Sequencing Reveals Differential Fibroblast Programming in the Presence or Absence of Epithelial Cells
On the basis of the capacity of the organoid system to rapidly establish epithelial repression of fibroblast activation, we sought to use it to begin the process of identifying key mediators of epithelial–mesenchymal reciprocal signaling and homeostasis. We therefore formed organoids from mTEs and, for the purpose of postorganoid sorting, fibroblasts isolated from Col1a1-GFP+ mice (CD31−/CD45−/CD326−/GFP+ cells). After culture of the mTEs and fibroblasts together, or of fibroblasts alone in identical organoid-supportive conditions, we harvested and resorted the GFP+ cells for analysis (Figure 5A). RNA was purified from these fibroblasts and analyzed by RNA sequencing (RNA-seq). RNA-seq demonstrated clear separation of gene expression between Col1a1-GFP+ fibroblasts grown alone and cocultured in organoids by principal component analysis (Figure 5B). Further analysis confirmed the dramatic transcriptional effects of the organoid coculture system with 1,992 differentially repressed genes and 2,156 differentially elevated genes in organoid-cultured fibroblasts compared with fibroblasts cultured alone (Table E1). To focus on characteristic fibroblast genes and the effects of organoid coculture, we selected for further analysis the top 1,200 most highly expressed transcripts in fibroblast-only cultures that were differentially expressed in organoids. Of these 1,200 transcripts, 802 were expressed at higher concentrations in fibroblasts alone, whereas 398 were expressed at higher concentrations in fibroblasts sorted from organoids (Figure 5C). Pathway analysis of these gene sets suggested that organoid coculture dramatically influenced metabolic programming of fibroblasts (Figure 5E). In contrast, fibroblasts cultured alone exhibited greater ECM, PI3K, and TNF pathway activation (Figure 5D). Among the transcripts differentially increased in fibroblasts alone were multiple genes implicated in fibroblast contractile activation (Acta2, Tagln [transgelin], Myl9 [myosin light chain 9]), ECM deposition (Fn1, Col1a2), ECM cross-linking (Loxl1, Loxl3), matrix metalloproteinase inhibition (Timp1 [tissue inhibitor of metalloproteinase inhibitor 1], Timp3), and profibrotic signaling (e.g., CTGF, Wnt4, Igf1 [insulin-like growth factor 1], Tgfbr1 [TGF-β receptor type 1], Pdgfrb [platelet-derived growth factor receptor-β]) (Figure 5F). Several potentially antifibrotic regulators were observed to be increased in organoid-cultured fibroblasts, including Klf4 [Kruppel-like factor 4] and Mfge8 [milk fat globule–epidermal growth factor 8] (42, 43). BMP signaling to fibroblasts in organoids also emerged as a potentially important regulatory mechanism for epithelial–mesenchymal cross-talk based on the significant increase in expression of Id1 (inhibitor of DNA binding 1) transcript in organoid-cultured fibroblasts relative to fibroblasts cultured alone (Figure 5F). Id1 is a characteristic transcriptional regulator and target downstream of BMP signaling (33) previously linked to inhibition of collagen production and fibrosis (44, 45).
Figure 5.
RNA sequencing (RNA-seq) reveals differential fibroblast programming in organoids. (A) Col1a1-GFP+ fibroblasts were isolated and cultured in organoid conditions alone or with mouse primary mTEs for 3 days, then organoids were harvested and Col1a1-GFP+ cells were isolated by FACS and subjected to RNA-seq (n = 3 per group). (B) Principal component (PC) analysis of RNA-seq results from organoid-cultured fibroblasts alone or cocultured with mTEs (n = 3 per group). (C) Heat map hierarchical clustering of genes and samples showing differentially expressed patterns of genes in organoid-cultured fibroblasts alone versus cocultured with mTEs. (D) Pathway analysis of upregulated gene sets in fibroblasts cultured alone. (E) Pathway analysis of upregulated gene sets in fibroblasts cocultured with mTEs. (F) Heat map of selected genes differentially expressed in organoid-cultured fibroblasts alone versus fibroblasts cocultured with mTEs showing fibroblast activation in the absence of mTEs. FDR = false discovery rate; HIF = hypoxia-inducible factor; MAX = maximum; MIN = minimum.
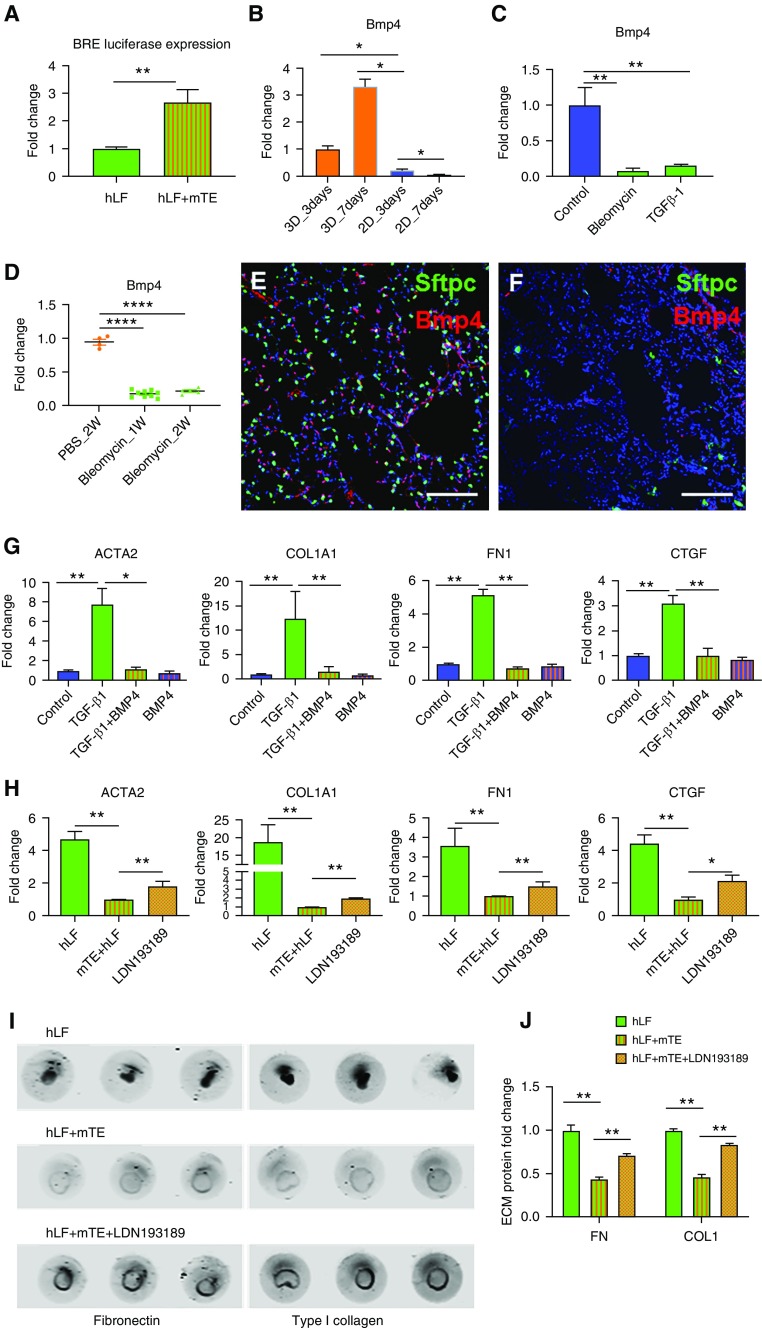
BMP Signaling Mediates Fibroblast Repression in Organoids
BMPs are known to play critical roles in tissue development, and specific BMPs such as BMP4 and BMP7 can act to counterbalance TGF-β signaling and exhibit antifibrogenic effects in multiple organs, including the kidney (46), lung (47), and liver (48). In lung development and maturation, BMP4 guides lung bud morphogenesis (49), regulates proximal–distal differentiation of endoderm (50), and promotes distal lung differentiation (51, 52), indicating critical roles in maintaining lung cellular phenotype and function. To evaluate the presence of functional BMP signaling in organoids, we transfected a BMP signaling reporter (pGL3 BRE Luciferase plasmid) into hLFs before organoid coculture, then quantified luciferase activity at 72 hours after organoid formation. BMP signaling activity was significantly higher in coculture organoids than in hLFs grown alone under the same culture conditions, confirming enhanced BMP signaling to fibroblasts in organoid coculture (Figure 6A). To directly assess Bmp4 transcript concentrations in lung epithelium, we used mouse Bmp4-specific primers to detect murine epithelial expression in RNA purified from organoids. Comparison of epithelial cells cultured alone in 2D culture with those cocultured in organoids with fibroblasts demonstrated that Bmp4 transcript concentrations were significantly higher as a result of the 3D organoid culture conditions (Figure 6B), consistent with the fibroblast reporter assay (Figure 6A), and suggested a potential role for Bmp4 signaling in epithelial repression of fibroblast activation. We also observed that Bmp4 transcript concentrations were significantly decreased with bleomycin or TGF-β1 treatment in vitro (Figure 6C), consistent with epithelial injury decreasing Bmp4 expression. To confirm the potential physiological relevance of Bmp4 in vivo as a repressed signal in physiological injury, we used immunofluorescence imaging to visualize Bmp4-positive cells in the lungs of mice expressing Sftpc-GFP. Representative images demonstrate the widespread expression of Bmp4 in the healthy adult mouse lung (Figure 6E), including within Sftpc-GFP+ cells, but also broad expression consistent with prior reports of Bmp4 expression in the lung microvasculature and stromal cells (53). To confirm epithelial Bmp4 expression in vivo, we isolated lung epithelial cells by flow sorting the CD326+/CD45−/CD31−/GFP− population from the Col1a1-GFP+ mouse and detected abundant Bmp4 transcripts in this cell population (Figure 6D). At 2 weeks after bleomycin exposure, global Bmp4 staining was nearly completely lost, consistent with prior work (47), as was the Sftpc-GFP signal (Figure 6F). Similarly, Bmp4 transcript concentrations were significantly reduced in freshly sorted CD326+ lung epithelial cells isolated after bleomycin exposure (Figure 6D).
Figure 6.
Bone morphogenetic protein (BMP) signaling partially mediates the epithelial antifibrotic effect. (A) BMP signaling activity in normal hLFs is shown by BRE luciferase assay in organoid-cultured fibroblasts alone and cocultured with mTEs. n = 6. (B) Comparison of Bmp4 expression in mTEs under organoid coculture conditions versus two-dimensional plastic culture conditions for 3 and 7 days. n = 4. (C) Comparison of Bmp4 expression in mTEs treated with bleomycin (15 μg/ml) or TGF-β1 (5 ng/ml). n = 6. (D) Gene expression analysis of Bmp4 in lung epithelial cells freshly sorted from PBS-treated (2 wk) and bleomycin-injured mice after 1 or 2 weeks. (E and F) Representative immunofluorescence images of Bmp4 in the lungs of PBS-treated (E) and bleomycin-treated (F) Sftpc-EGFP mice at 2 weeks. Scale bar: 100 μm. (G) Fibroblast-specific expression of ACTA2, COL1A1, FN1, and CTGF in three-dimensional (3D) coculture organoids treated with TGF-β1 (5 ng/ml) only, TGF-β1 (5 ng/ml) plus BMP4 (10 ng/ml), or BMP4 only (10 ng/ml) for 48 hours compared with no-treatment control. n = 6. (H) Increased fibroblast-specific expression of ACTA2, COL1A1, FN1, and CTGF after LDN193189 (1 μM) treatment for 48 hours in 3D coculture organoids. n = 6. (I) Reversal of reduced ECM accumulation of FN and collagen I (COL1) in hLF + mTE organoids by the BMP inhibitor LDN193189. (J) Quantification of FN and COL1 in hLFs alone or hLF + mTE cocultures with or without LDN193189. n = 5. *P < 0.05, **P < 0.01, and ****P < 0.0001.
To test the functional effects of BMP4 signaling within organoids, we treated coculture organoids with TGF-β1 and BMP4 alone or in combination. As expected, TGF-β1 treatment increased fibroblast expression of ACTA2, COL1A1, FN1, and CTGF when applied to organoids as a whole (Figure 6G), consistent with our previously published results (29). Cotreatment of organoids with BMP4 strongly attenuated the expression of all four genes induced by TGF-β1 (Figure 6G), in agreement with prior observations (54). In contrast, BMP4 treatment alone did not significantly reduce baseline expression of these genes in organoids (Figure 6G). This result is consistent with the already high levels of endogenous Bmp4 expression and signaling noted above. Furthermore, the efficacy of exogenous BMP4 in TGF-β1–stimulated organoids is consistent with the emergence of a BMP4-responsive window initiated by the TGF-β1–mediated reduction in epithelial Bmp4 expression that we previously observed (Figure 6C).
The findings described above suggest that constitutive BMP signaling may contribute to the baseline repression of fibroblast activation in organoid cocultures. To test this concept, we used a BMP signaling antagonist, LDN193189 (LDN) (55), to inhibit endogenous BMP signaling. Treatment with LDN significantly, though incompletely, attenuated the decrease in fibroblast gene expression induced by the interaction between coculture organoid populations (Figure 6H). At the ECM deposition level, treatment with LDN significantly increased fibronectin and type I collagen deposition (Figures 6I and 6J), largely reversing the repressive effects provided by the cocultured epithelial cells in organoids. Taken together, these data demonstrate that endogenous BMP signaling plays an important role in epithelial repression of the fibroblast activation state in the organoid cocultures. Our observations that Bmp4 expression is lost with epithelial injury and that exogenous BMP4 reduces fibroblast activation in these settings mirror similar observations made in mouse models of lung injury and fibrosis (47, 56), but for the first time they reveal how epithelial injury alone is coupled to fibroblast activation through loss of fibroblast-repressive BMP signaling. Thus, this relatively simple epithelial–mesenchymal nascent organoid culture system appears capable of replicating key cell–cell interactions present during homeostasis, injury, and fibrosis in vivo, and it should provide a powerful platform on which to discover and dissect additional candidate pathways and mediators of epithelial–mesenchymal homeostasis and activation.
Discussion
Epithelial–mesenchymal interactions are pivotal during lung development (3, 4, 13), and their pathological interactions are believed to underlie airway and alveolar fibrosis in diseases such asthma and IPF (6, 7, 10, 11, 41). Complex cell–cell interactions are challenging to disentangle using in vivo systems, and organoids thus represent an important new tool through which to interrogate epithelial–mesenchymal interactions relevant to health and disease. Although prior work using organoids was largely focused on mesenchymal influence on the epithelial state, our present work emphasizes the important reciprocal effects of epithelium on mesenchyme, demonstrating that such effects represent an important restraining effect on fibroblasts that may be lost during injury or epithelial dysfunction, leading to fibroblast activation and fibrotic matrix deposition. Specifically, our results demonstrate that healthy proximal or distal epithelial cells dramatically repress transcriptional and functional metrics of contractile and fibrogenic fibroblast activation, both at baseline and after fibroblast-only TGF-β1 stimulation. In contrast, epithelial injury before organoid culture, by either bleomycin or TGF-β1 exposure, is sufficient alone to promote fibroblast activation. By comparing bleomycin and TGF-β1 responses, we highlight that the default common response to epithelial perturbation, whether by bleomycin injury or by TGF-β1 stimulation, is to reduce repressive effects on fibroblast activation. Although previous work has largely ascribed such effects to epithelium-derived profibrotic factors, our work demonstrates that loss of epithelium-derived fibroblast-repressive factors likely also plays an important role, with Bmp4 identified as a relevant factor that represses fibroblast activation and is lost after epithelial injury in vivo and in vitro. Importantly, constitutive Bmp4 expression and Bmp signaling have also recently been described as important brakes on proximal and distal epithelial proliferation, and their transient loss is critical to epithelial regeneration after injury (52, 57, 58). Hence, our findings provide a novel link whereby lung injury is inherently coupled to coordinated mesenchymal activation and epithelial regeneration via repressed Bmp signaling. Moreover, accumulating evidence implicates persistent repression of Bmp signaling in fibrotic pathologies, suggesting that failure to restore this homeostatic relationship may promote persistent fibroblast activation and fibrosis (47, 56, 59–62).
It has long been appreciated that the complexity of lung biology requires taking into account multiple cell types and their interactions in both health and disease (4, 6, 7, 13, 20). Prior coculture studies have predominantly focused on monolayer coculture or coculture Transwell assay (12, 63–67). Although these studies have advanced understanding of epithelial–mesenchymal interactions, they have some limitations. 2D monolayers of cells cultured on flat and rigid substrates are much stiffer than soft tissues such as the lung, which may artificially stimulate cellular activation of mesenchymal (68, 69) and epithelial (70) cell types. Such systems also limit the exposure of cells to close cell–cell interactions, particularly through cell–cell contact and a shared ECM environment (71). Organoid culture systems address these limitations, allowing for a more physiologic setting for close cell–cell interactions and 3D matrix and morphogenetic remodeling. An interesting aspect of our study is that we found no significant effect of epithelium on profibrotic gene expression in a traditional 2D culture system, but we demonstrated significant differences in our organoid system (Figures 1C and 1D). Of note, we also found that mouse epithelial cells express higher concentrations of Bmp4 in organoids than in monolayer culture and that Bmp4 is abundantly expressed in healthy mouse lung in vivo, proving one example of how organoid culture conditions induce changes in expression of an important paracrine mediator to provide a physiologic context for the study of epithelial–mesenchymal interactions.
Epithelial–mesenchymal cross-talk is essential throughout development, from lung branching to alveologenesis (3, 4, 13, 72), and dysfunctional epithelial–mesenchymal interactions lead to fibrotic diseases of both the proximal airways and the distal gas exchange regions of the lung (5–7, 10, 11, 41). A notable finding of our work is that epithelial cells from both proximal (tracheal and bronchial) and distal (SpC+) locations exhibit profound repressive effects on fibroblast activation. Our findings are consistent with the observation that conditional deletion of epithelial populations in either the airways or the distal lung alone is sufficient to generate a local fibrotic response (73, 74). Several open questions remain regarding the nature of this relationship between epithelium and mesenchyme. For example, do proximal and distal epithelial cells confer this repressive effect on mesenchyme through shared or distinct signaling pathways? Because the loss of such repressive signals may play important roles in fibrotic remodeling of the proximal airways as well as distal lung, defining such interactions takes on elevated relevance. Moreover, it has been shown that fibroblast populations from proximal and distal lung sites exhibit different markers and behaviors (24, 75). An additional open question not addressed by our work is whether fibroblasts selectively isolated from these locations exhibit differential responses to proximal and distal epithelium and epithelial injury. The methods developed in the present study should allow for detailed investigation of such important issues. Future refinement of our approach, with inclusion of human disease–derived cells as well as additional important lung cell types such as endothelium and macrophages (51, 76, 77), may allow even greater insight into the cell–cell interactions that underlie homeostasis and are perturbed in chronic diseases of the lung.
Although significant challenges remain in gaining a full understanding of the mechanisms of epithelial–mesenchymal cross-talk in normal lung homeostasis and lung disease development, our results emphasize an important and relatively understudied concept that healthy epithelial cells engage in continuous interactions with neighboring mesenchyme to sustain their relatively quiescent state of low contractile proliferative and matrix synthetic activity. How epithelial cell injury activates fibroblasts has been studied extensively for many years, with a predominant focus on identifying fibrogenic activators released by injured epithelium in vitro and in vivo (5–7, 10, 11, 41). In this study, we show that healthy epithelial cells also suppress the profibrotic activation of human fibroblasts at baseline and after fibroblast stimulation by TGF-β1, demonstrating an active epithelial role in this process. Epithelial perturbation alone is sufficient to impair this repressive function, likely reflecting not only the release of profibrotic factors but also reduced signaling through fibroblast-repressive signals such as BMP4 as identified in the present study, as well as previously described factors such as prostaglandin E2 (78, 79). Importantly, this new appreciation for healthy epithelial repression implies that fibrogenesis may proceed whenever epithelial function is perturbed (73, 74) and does not necessarily require epithelium-derived profibrotic factors. Hence, additional efforts to identify, and ultimately restore, epithelium-derived fibroblast-repressive signals that are lost in disease may provide a fruitful avenue for discovery and therapeutic intervention.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Dr. Derek Radisky for providing Col1a1-GFP transgenic mice. The authors also thank Bruce W. Eckloff and Daniel O’Brien for RNA-seq data acquisition and analysis. The authors appreciate the generous access to equipment supported by Dr. Y. S. Prakash and Dr. Gary C. Sieck.
Footnotes
Supported by National Institutes of Health grants R01 HL092961 (D.J.T.), R21 HL132256 (D.J.T.), R01 HL142596 (G.L.), and HL105355 (D.L.J.).
Author Contributions: Q.T.: conceptualization, data curation, formal analysis, supervision, investigation, methodology, writing of the original draft, project administration, manuscript review and editing. X.Y.M.: investigation, data curation, formal analysis. W.L.: investigation, data curation. J.A.M.: data curation, methodology. D.L.J.: methodology. A.J.H.: methodology. D.S.: data curation, methodology. G.L.: funding acquisition, methodology, manuscript review and editing. D.J.T.: funding acquisition, conceptualization, formal analysis, supervision, investigation, methodology, writing of the original draft, project administration, and manuscript review and editing.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0390OC on May 3, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 3.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shannon JM, Hyatt BA. Epithelial–mesenchymal interactions in the developing lung. Annu Rev Physiol. 2004;66:625–645. doi: 10.1146/annurev.physiol.66.032102.135749. [DOI] [PubMed] [Google Scholar]
- 5.Chapman HA. Epithelial–mesenchymal interactions in pulmonary fibrosis. Annu Rev Physiol. 2011;73:413–435. doi: 10.1146/annurev-physiol-012110-142225. [DOI] [PubMed] [Google Scholar]
- 6.Horowitz JC, Thannickal VJ. Epithelial–mesenchymal interactions in pulmonary fibrosis. Semin Respir Crit Care Med. 2006;27:600–612. doi: 10.1055/s-2006-957332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holgate ST, Davies DE, Lackie PM, Wilson SJ, Puddicombe SM, Lordan JL. Epithelial–mesenchymal interactions in the pathogenesis of asthma. J Allergy Clin Immunol. 2000;105:193–204. doi: 10.1016/s0091-6749(00)90066-6. [DOI] [PubMed] [Google Scholar]
- 8.Thannickal VJ, Toews GB, White ES, Lynch JP, III, Martinez FJ. Mechanisms of pulmonary fibrosis. Annu Rev Med. 2004;55:395–417. doi: 10.1146/annurev.med.55.091902.103810. [DOI] [PubMed] [Google Scholar]
- 9.Selman M, Pardo A. Idiopathic pulmonary fibrosis: an epithelial/fibroblastic cross-talk disorder. Respir Res. 2002;3:3. doi: 10.1186/rr175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hewlett JC, Kropski JA, Blackwell TS. Idiopathic pulmonary fibrosis: epithelial–mesenchymal interactions and emerging therapeutic targets. Matrix Biol. 2018;71–72:112–127. doi: 10.1016/j.matbio.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies DE. The role of the epithelium in airway remodeling in asthma. Proc Am Thorac Soc. 2009;6:678–682. doi: 10.1513/pats.200907-067DP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osei ET, Noordhoek JA, Hackett TL, Spanjer AI, Postma DS, Timens W, et al. Interleukin-1α drives the dysfunctional cross-talk of the airway epithelium and lung fibroblasts in COPD. Eur Respir J. 2016;48:359–369. doi: 10.1183/13993003.01911-2015. [DOI] [PubMed] [Google Scholar]
- 13.Hines EA, Sun X. Tissue crosstalk in lung development. J Cell Biochem. 2014;115:1469–1477. doi: 10.1002/jcb.24811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Invest. 2012;122:2756–2762. doi: 10.1172/JCI60323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng T, Frank DB, Kadzik RS, Morley MP, Rathi KS, Wang T, et al. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature. 2015;526:578–582. doi: 10.1038/nature14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Smartt H, Holgate ST, Roche WR. Growth factors secreted by bronchial epithelial cells control myofibroblast proliferation: an in vitro co-culture model of airway remodeling in asthma. Lab Invest. 1999;79:395–405. [PubMed] [Google Scholar]
- 17.Ishikawa S, Ishimori K, Ito S. A 3D epithelial–mesenchymal co-culture model of human bronchial tissue recapitulates multiple features of airway tissue remodeling by TGF-β1 treatment. Respir Res. 2017;18:195. doi: 10.1186/s12931-017-0680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osei ET, Florez-Sampedro L, Tasena H, Faiz A, Noordhoek JA, Timens W, et al. miR-146a-5p plays an essential role in the aberrant epithelial-fibroblast cross-talk in COPD. Eur Respir J. 2017;49:1602538. doi: 10.1183/13993003.02538-2016. [DOI] [PubMed] [Google Scholar]
- 19.Suwara MI, Green NJ, Borthwick LA, Mann J, Mayer-Barber KD, Barron L, et al. IL-1α released from damaged epithelial cells is sufficient and essential to trigger inflammatory responses in human lung fibroblasts. Mucosal Immunol. 2014;7:684–693. doi: 10.1038/mi.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller AJ, Spence JR. In vitro models to study human lung development, disease and homeostasis. Physiology (Bethesda) 2017;32:246–260. doi: 10.1152/physiol.00041.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, et al. In vitro generation of human pluripotent stem cell derived lung organoids Elife 20154e05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegab AE, Arai D, Gao J, Kuroda A, Yasuda H, Ishii M, et al. Mimicking the niche of lung epithelial stem cells and characterization of several effectors of their in vitro behavior. Stem Cell Res (Amst) 2015;15:109–121. doi: 10.1016/j.scr.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, et al. Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell. 2017;170:1149–1163.e12. doi: 10.1016/j.cell.2017.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148.e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McQualter JL, Yuen K, Williams B, Bertoncello I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci USA. 2010;107:1414–1419. doi: 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikolić MZ, Caritg O, Jeng Q, Johnson JA, Sun D, Howell KJ, et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. Elife. 2017;6:e26575. doi: 10.7554/eLife.26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan Q, Choi KM, Sicard D, Tschumperlin DJ. Human airway organoid engineering as a step toward lung regeneration and disease modeling. Biomaterials. 2017;113:118–132. doi: 10.1016/j.biomaterials.2016.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yata Y, Scanga A, Gillan A, Yang L, Reif S, Breindl M, et al. DNase I-hypersensitive sites enhance α1(I) collagen gene expression in hepatic stellate cells. Hepatology. 2003;37:267–276. doi: 10.1053/jhep.2003.50067. [DOI] [PubMed] [Google Scholar]
- 31.Oh RS, Haak AJ, Smith KMJ, Ligresti G, Choi KM, Xie T, et al. RNAi screening identifies a mechanosensitive ROCK-JAK2-STAT3 network central to myofibroblast activation. J Cell Sci. 2018;131:jcs209932. doi: 10.1242/jcs.209932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanderbilt JN, Gonzalez RF, Allen L, Gillespie A, Leaffer D, Dean WB, et al. High-efficiency type II cell-enhanced green fluorescent protein expression facilitates cellular identification, tracking, and isolation. Am J Respir Cell Mol Biol. 2015;53:14–21. doi: 10.1165/rcmb.2014-0348MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 34.Kendall RT, Feghali-Bostwick CA. Fibroblasts in fibrosis: novel roles and mediators. Front Pharmacol. 2014;5:123. doi: 10.3389/fphar.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 36.Takebe T, Enomura M, Yoshizawa E, Kimura M, Koike H, Ueno Y, et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell. 2015;16:556–565. doi: 10.1016/j.stem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Mio T, Liu XD, Adachi Y, Striz I, Sköld CM, Romberger DJ, et al. Human bronchial epithelial cells modulate collagen gel contraction by fibroblasts. Am J Physiol. 1998;274:L119–L126. doi: 10.1152/ajplung.1998.274.1.L119. [DOI] [PubMed] [Google Scholar]
- 38.Hoyles RK, Derrett-Smith EC, Khan K, Shiwen X, Howat SL, Wells AU, et al. An essential role for resident fibroblasts in experimental lung fibrosis is defined by lineage-specific deletion of high-affinity type II transforming growth factor β receptor. Am J Respir Crit Care Med. 2011;183:249–261. doi: 10.1164/rccm.201002-0279OC. [DOI] [PubMed] [Google Scholar]
- 39.Li M, Krishnaveni MS, Li C, Zhou B, Xing Y, Banfalvi A, et al. Epithelium-specific deletion of TGF-β receptor type II protects mice from bleomycin-induced pulmonary fibrosis. J Clin Invest. 2011;121:277–287. doi: 10.1172/JCI42090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moeller A, Ask K, Warburton D, Gauldie J, Kolb M. The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol. 2008;40:362–382. doi: 10.1016/j.biocel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai N, Tager AM. Fibrosis of two: epithelial cell-fibroblast interactions in pulmonary fibrosis. Biochim Biophys Acta. 2013;1832:911–921. doi: 10.1016/j.bbadis.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atabai K, Jame S, Azhar N, Kuo A, Lam M, McKleroy W, et al. Mfge8 diminishes the severity of tissue fibrosis in mice by binding and targeting collagen for uptake by macrophages. J Clin Invest. 2009;119:3713–3722. doi: 10.1172/JCI40053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin L, Han Q, Xiong Y, Li T, Liu Z, Xu H, et al. Krüpple-like-factor 4 attenuates lung fibrosis via inhibiting epithelial–mesenchymal transition. Sci Rep. 2017;7:15847. doi: 10.1038/s41598-017-14602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Je YJ, Choi DK, Sohn KC, Kim HR, Im M, Lee Y, et al. Inhibitory role of Id1 on TGF-β-induced collagen expression in human dermal fibroblasts. Biochem Biophys Res Commun. 2014;444:81–85. doi: 10.1016/j.bbrc.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Lin L, Zhou Z, Zheng L, Alber S, Watkins S, Ray P, et al. Cross talk between Id1 and its interactive protein Dril1 mediate fibroblast responses to transforming growth factor-β in pulmonary fibrosis. Am J Pathol. 2008;173:337–346. doi: 10.2353/ajpath.2008.070915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morrissey J, Hruska K, Guo G, Wang S, Chen Q, Klahr S. Bone morphogenetic protein-7 improves renal fibrosis and accelerates the return of renal function. J Am Soc Nephrol. 2002;13:S14–S21. [PubMed] [Google Scholar]
- 47.De Langhe E, Cailotto F, De Vooght V, Aznar-Lopez C, Vanoirbeek JA, Luyten FP, et al. Enhanced endogenous bone morphogenetic protein signaling protects against bleomycin induced pulmonary fibrosis. Respir Res. 2015;16:38. doi: 10.1186/s12931-015-0202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang LP, Dong JZ, Xiong LJ, Shi KQ, Zou ZL, Zhang SN, et al. BMP-7 attenuates liver fibrosis via regulation of epidermal growth factor receptor. Int J Clin Exp Pathol. 2014;7:3537–3547. [PMC free article] [PubMed] [Google Scholar]
- 49.Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- 50.Weaver M, Yingling JM, Dunn NR, Bellusci S, Hogan BL. Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development. 1999;126:4005–4015. doi: 10.1242/dev.126.18.4005. [DOI] [PubMed] [Google Scholar]
- 51.Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung MI, Bujnis M, Barkauskas CE, Kobayashi Y, Hogan BLM. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 2018;145:dev163014. doi: 10.1242/dev.163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dyer LA, Pi X, Patterson C. The role of BMPs in endothelial cell function and dysfunction. Trends Endocrinol Metab. 2014;25:472–480. doi: 10.1016/j.tem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pegorier S, Campbell GA, Kay AB, Lloyd CM. Bone morphogenetic protein (BMP)-4 and BMP-7 regulate differentially transforming growth factor (TGF)-β1 in normal human lung fibroblasts (NHLF) Respir Res. 2010;11:85. doi: 10.1186/1465-9921-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong CC, Yu PB. Applications of small molecule BMP inhibitors in physiology and disease. Cytokine Growth Factor Rev. 2009;20:409–418. doi: 10.1016/j.cytogfr.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koli K, Myllärniemi M, Vuorinen K, Salmenkivi K, Ryynänen MJ, Kinnula VL, et al. Bone morphogenetic protein-4 inhibitor gremlin is overexpressed in idiopathic pulmonary fibrosis. Am J Pathol. 2006;169:61–71. doi: 10.2353/ajpath.2006.051263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, et al. Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell. 2016;19:217–231. doi: 10.1016/j.stem.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tadokoro T, Gao X, Hong CC, Hotten D, Hogan BL. BMP signaling and cellular dynamics during regeneration of airway epithelium from basal progenitors. Development. 2016;143:764–773. doi: 10.1242/dev.126656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong Y, Geng Y, Li L, Li X, Yan X, Fang Y, et al. Blocking follistatin-like 1 attenuates bleomycin-induced pulmonary fibrosis in mice. J Exp Med. 2015;212:235–252. doi: 10.1084/jem.20121878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farkas L, Farkas D, Gauldie J, Warburton D, Shi W, Kolb M. Transient overexpression of Gremlin results in epithelial activation and reversible fibrosis in rat lungs. Am J Respir Cell Mol Biol. 2011;44:870–878. doi: 10.1165/rcmb.2010-0070OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy N, Gaynor KU, Rowan SC, Walsh SM, Fabre A, Boylan J, et al. Altered expression of bone morphogenetic protein accessory proteins in murine and human pulmonary fibrosis. Am J Pathol. 2016;186:600–615. doi: 10.1016/j.ajpath.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 62.Myllärniemi M, Lindholm P, Ryynänen MJ, Kliment CR, Salmenkivi K, Keski-Oja J, et al. Gremlin-mediated decrease in bone morphogenetic protein signaling promotes pulmonary fibrosis. Am J Respir Crit Care Med. 2008;177:321–329. doi: 10.1164/rccm.200706-945OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mangum JB, Everitt JI, Bonner JC, Moore LR, Brody AR. Co-culture of primary pulmonary cells to model alveolar injury and translocation of proteins. In Vitro Cell Dev Biol. 1990;26:1135–1143. doi: 10.1007/BF02623690. [DOI] [PubMed] [Google Scholar]
- 64.Alfaro-Moreno E, Nawrot TS, Vanaudenaerde BM, Hoylaerts MF, Vanoirbeek JA, Nemery B, et al. Co-cultures of multiple cell types mimic pulmonary cell communication in response to urban PM10. Eur Respir J. 2008;32:1184–1194. doi: 10.1183/09031936.00044008. [DOI] [PubMed] [Google Scholar]
- 65.Hermanns MI, Unger RE, Kehe K, Peters K, Kirkpatrick CJ. Lung epithelial cell lines in coculture with human pulmonary microvascular endothelial cells: development of an alveolo-capillary barrier in vitro. Lab Invest. 2004;84:736–752. doi: 10.1038/labinvest.3700081. [DOI] [PubMed] [Google Scholar]
- 66.Greer RM, Miller JD, Okoh VO, Halloran BA, Prince LS. Epithelial–mesenchymal co-culture model for studying alveolar morphogenesis. Organogenesis. 2014;10:340–349. doi: 10.4161/org.29198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swartz MA, Tschumperlin DJ, Kamm RD, Drazen JM. Mechanical stress is communicated between different cell types to elicit matrix remodeling. Proc Natl Acad Sci USA. 2001;98:6180–6185. doi: 10.1073/pnas.111133298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wells RG. The role of matrix stiffness in regulating cell behavior. Hepatology. 2008;47:1394–1400. doi: 10.1002/hep.22193. [DOI] [PubMed] [Google Scholar]
- 69.Liu F, Lagares D, Choi KM, Stopfer L, Marinković A, Vrbanac V, et al. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2015;308:L344–L357. doi: 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, et al. Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat Cell Biol. 2015;17:678–688. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bertoncello I. Stem cells in the lung: development, repair and regeneration. New York: Springer; 2015. [Google Scholar]
- 73.Perl AK, Riethmacher D, Whitsett JA. Conditional depletion of airway progenitor cells induces peribronchiolar fibrosis. Am J Respir Crit Care Med. 2011;183:511–521. doi: 10.1164/rccm.201005-0744OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sisson TH, Mendez M, Choi K, Subbotina N, Courey A, Cunningham A, et al. Targeted injury of type II alveolar epithelial cells induces pulmonary fibrosis. Am J Respir Crit Care Med. 2010;181:254–263. doi: 10.1164/rccm.200810-1615OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou X, Wu W, Hu H, Milosevic J, Konishi K, Kaminski N, et al. Genomic differences distinguish the myofibroblast phenotype of distal lung fibroblasts from airway fibroblasts. Am J Respir Cell Mol Biol. 2011;45:1256–1262. doi: 10.1165/rcmb.2011-0065OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X, Franklin RA, Adler M, Jacox JB, Bailis W, Shyer JA, et al. Circuit design features of a stable two-cell system. Cell. 2018;172:744–757.e17. doi: 10.1016/j.cell.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lechner AJ, Driver IH, Lee J, Conroy CM, Nagle A, Locksley RM, et al. Recruited monocytes and type 2 immunity promote lung regeneration following pneumonectomy. Cell Stem Cell. 2017;21:120–134.e7. doi: 10.1016/j.stem.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bozyk PD, Moore BB. Prostaglandin E2 and the pathogenesis of pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:445–452. doi: 10.1165/rcmb.2011-0025RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Epa AP, Thatcher TH, Pollock SJ, Wahl LA, Lyda E, Kottmann RM, et al. Normal human lung epithelial cells inhibit transforming growth factor-β induced myofibroblast differentiation via prostaglandin E2. PLoS One. 2015;10:e0135266. doi: 10.1371/journal.pone.0135266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.