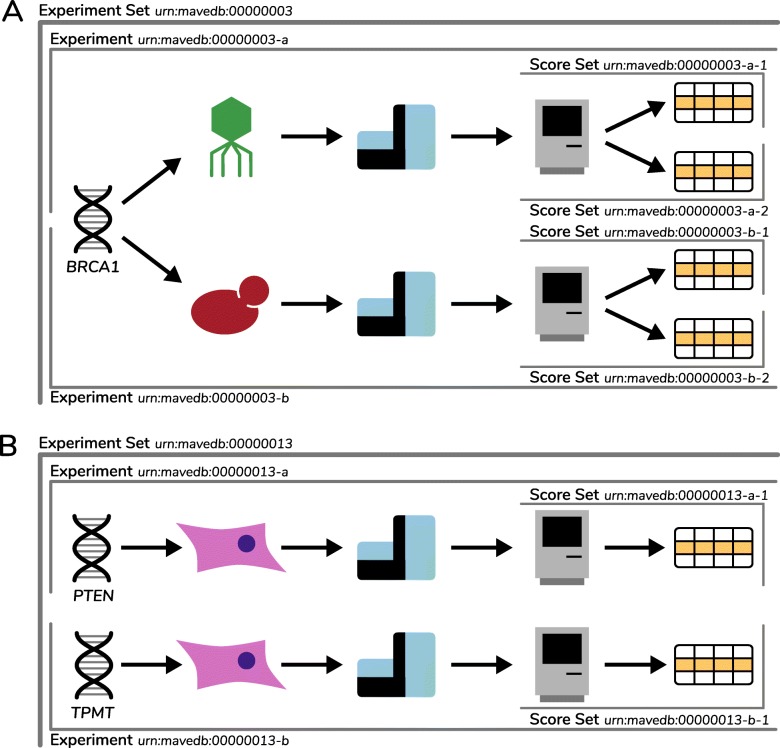
Fig. 1.
Representative structures of MaveDB entries. Each panel depicts a single experiment set and its associated accession numbers. a A single target sequence analyzed using two different MAVEs. For example, MAVEs in phage display and yeast two-hybrid formats were performed on BRCA1 (experiment accessions ending in “-a” and “-b”) [16]. Each of these yielded two score sets, one for amino acid variants and one for nucleotide variants (score set accessions ending in “-1” and “-2”). b Two distinct target sequences analyzed using the same MAVE. For example, the VAMP-seq assay was performed on the PTEN and TPMT target sequences (experiment accessions ending in “-a” and “-b”) [18], yielding an amino acid score set for each target (score set accessions ending in “-1”)