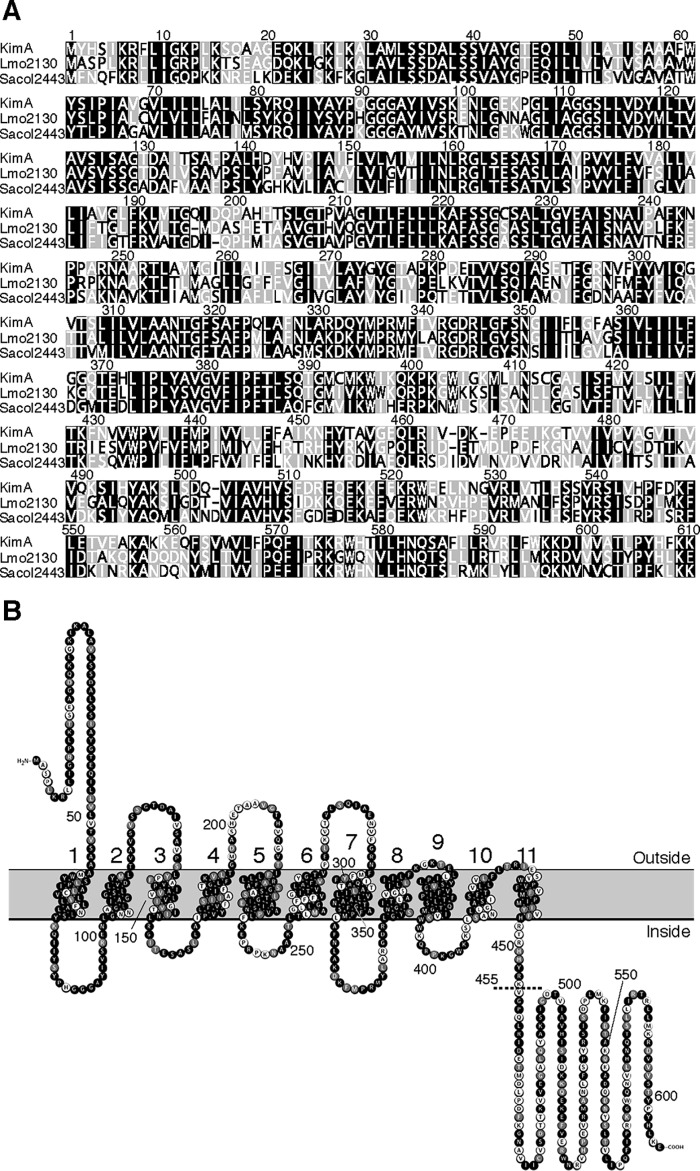
Figure 1.
Alignment of KimA homologs and domain organization of the KimALmo protein. A, MUSCLE alignment of the KimABsu, KimALmo, and KimASau homologs from B. subtilis, L. monocytogenes (Lmo2130), and S. aureus (Sacol2443), respectively, generated with the Geneious software package (63). Amino acids in black, gray, and white have an amino acid similarity of >80, 60–80, or <60%, respectively. B, predicted membrane topology of KimALmo overlaid with a MUSCLE alignment between KimABsu and KimALmo. The dashed line indicates the position at which the KimALmo protein was truncated. Amino acids in black are identical; amino acids in gray are similar, and amino acids in white are nonsimilar.