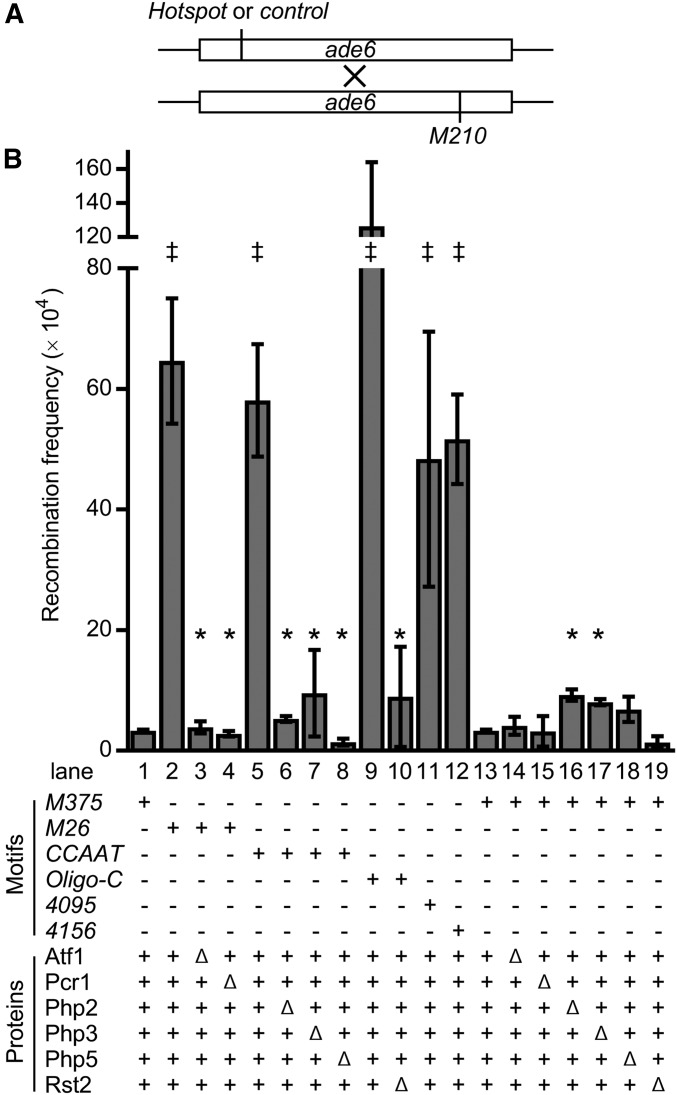
Figure 1.
DNA sequence-specific, binding protein-dependent activation of meiotic recombination hotspots. (A) Assay for meiotic recombination. Diagram shows ade6 ORF (boxes) and relative positions of alleles used. Haploid cells harboring a basal recombination control allele (M375) or alleles that contain a hotspot DNA sequence motif (M26, CCAAT, Oligo-C, 4095, and 4156) were crossed to a strain with a tester allele (M210) and haploid meiotic products were scored for frequencies of ade6+ recombinants. (B) Recombinant frequencies from basal control and hotspot crosses in the presence or absence of proteins that bind to the hotspot DNA sequence motifs. Data are mean ± SD from three biological replicates; statistically significant differences (P ≤ 0.05% from t-test) are shown for hotspot vs. control (‡) and hotspot lacking its binding proteins vs. with its binding proteins (*). Note that hotspot activation requires both the DNA sequence motif and the protein(s) that bind to that motif, and that ablating these binding proteins does not reduce significantly rates of basal recombination at the control M375. The DNA sequences and precise locations of alleles are provided in Table S2; primary data values are provided in Table S4.