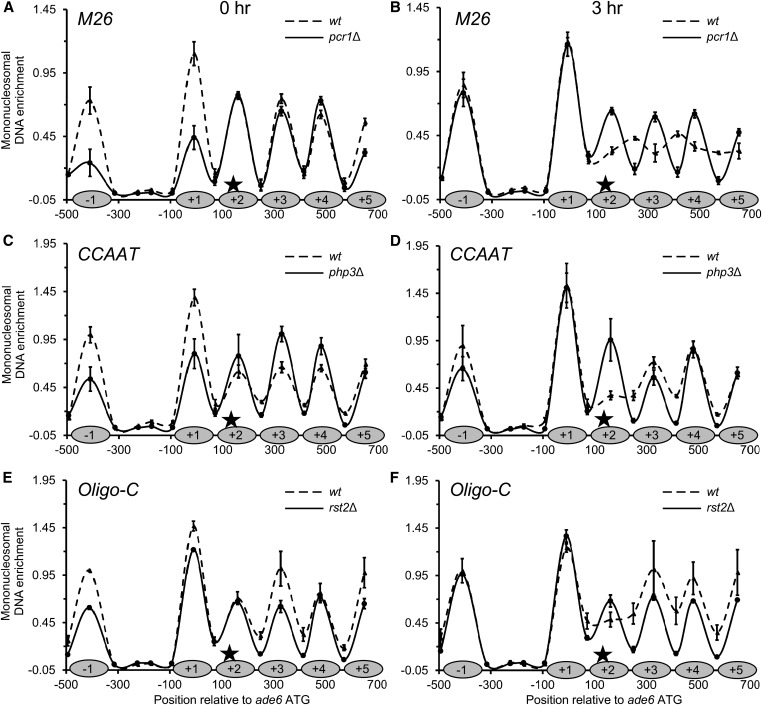
Figure 4.
DNA sequence-specific binding proteins regulate chromatin remodeling at sequence-dependent hotspots. Chromatin structures of hotspot alleles before (0 hr; A, C, and E) and during meiosis (3 hr; B, D, and F) in wild-type cells and in cells lacking the respective DNA binding proteins. Data are mean ± SD from three biological replicates, with internal normalization to a nucleosome in smc5. The positions of nucleosomes in the basal recombination control [from (A) of Figure 3] are depicted on each X-axis (shaded ovals) for the sake of comparison. Note that ablating the hotspot-activating binding proteins restores a more normal phasing of nucleosomes and that this effect is most pronounced at 3 hr of meiosis. Tabular data used to generate this figure are provided in Table S5.