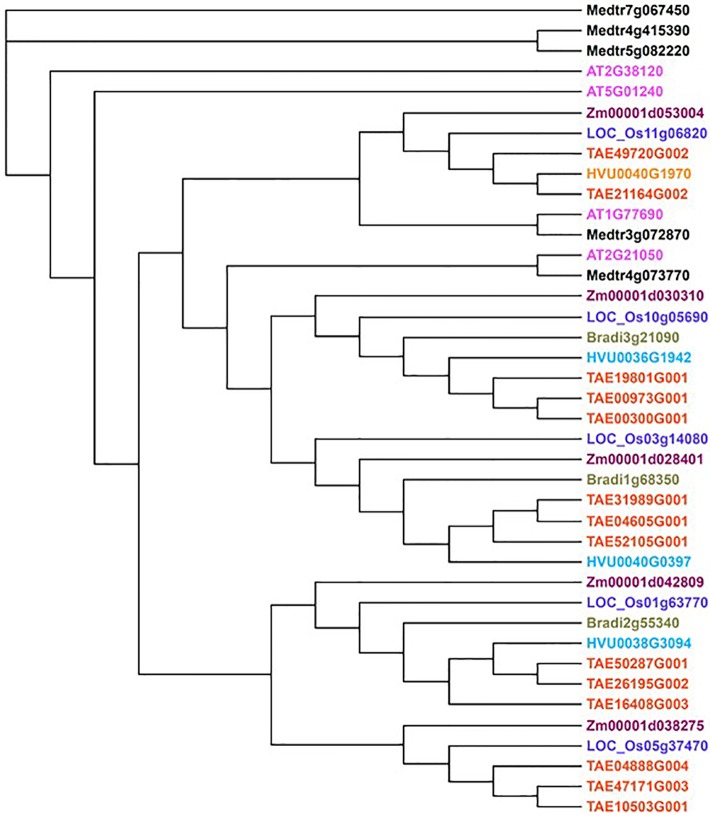
Figure 2.
The phylogeny of AUX1 homologs of selected plant species. This tree was generated using interactive phylogenetic module from Plaza 4.0 (Van Bel et al., 2017). AtAUX1 was used as seed gene to retrieve homologs of selected species [Arabisdopsis thaliana (AT), Zea maize (Zm), Oryza sativa ssp. Japonica (LOC), Triticum aestivum (TAE), Hordeum vulgare (HVU), Brachypodium distachyon (Bradi) and Medicago truncatula (Medtr); coloured differently] and homologs with >70% protein identity were retained for generating the tree. Some of these homologs have been previously characterised (discussed in the main text).