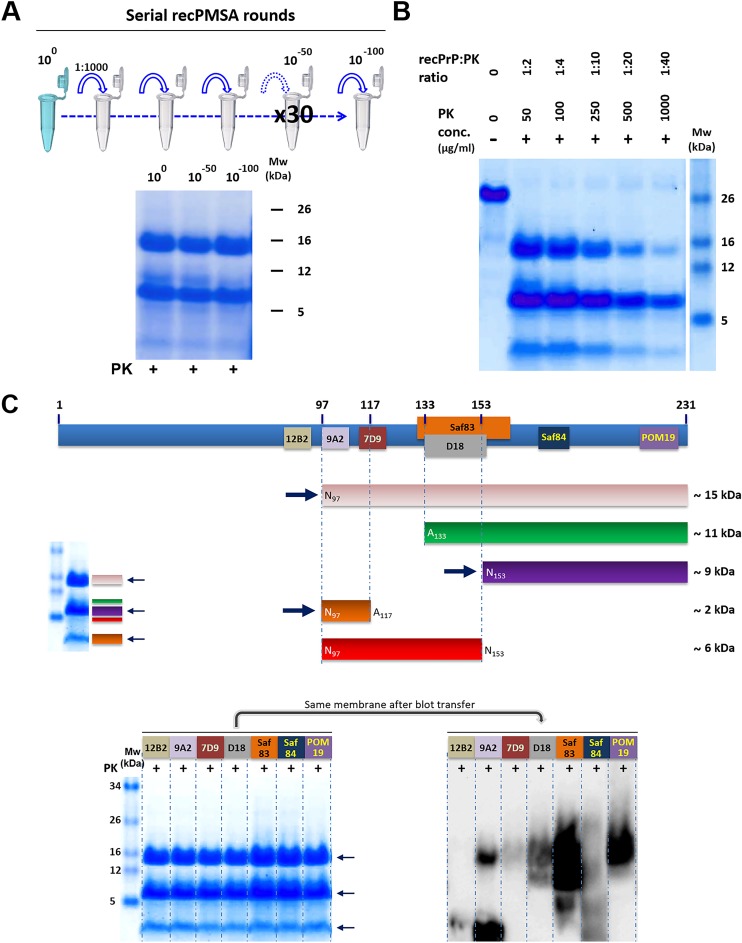
Fig 7. Biochemical characterization of L-seeded-PMSA.
A) The ability of L-seeded-PMSA to self-propagate indefinitely shown through 30 serial rounds of PMSA at 1:1,000 dilution. 500 μl of the original seed (100 dilution), the product of round 17 of PMSA (10−50 dilution) and the product of round 33 (10−100 dilution) were PK-digested, concentrated by centrifugation and visualized by total protein staining and showed no detectable pattern differences throughout the propagation process. B) Protease-K (PK) resistance of L-seeded PMSA was evaluated by submitting fractions of the same sample to increasing PK concentrations for 1h at 42°C. L-seeded-PMSA showed high resistance to protease, as the main PK-resistant bands are maintained even at the highest PK concentration used. C) Identification of the proteolytic fragments of L-seeded-PMSA by epitope mapping was performed with the same PK-digested and concentrated sample divided in seven that were stained for total protein to guarantee equal protein amounts in each lane. The stained gel was then transferred for Western blotting and the following antibodies were used for epitope mapping: 12B2 (1:2,500), 9A2 (1:4,000), 7D9 (1:1,000), D18 (1:5,000), Saf83 (1:400), Sa84 (1:400) and POM19 (1:10,000). The theoretical reconstruction of the PrP fragments recognized by each antibody are represented in the cartoon, the most abundant fragments were (see arrows) ~15 (pink), ~9 (purple) and ~2 (orange) kDa. Another two minor bands reflect other PK cleavages yielding two fragments of ~12 and ~6 kDa (green and red bands, respectively). These fragments are not always detected by all the antibodies used for the epitope mapping due to their low amounts, below the detection limit of some of the antibodies. MW: Molecular weight.