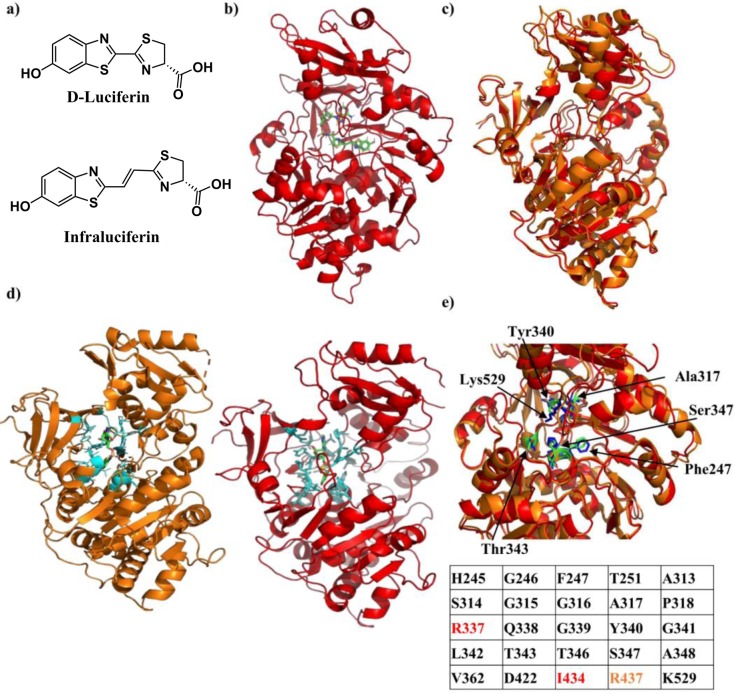
Figure 1. Crystal structure of Firefly luciferase in complex with a iLH2 analogue.
(a) Chemical structures of native D-Luciferin (LH2) and the LH2 analogue infraluciferin (iLH2). (b) The crystal structure of Firefly luciferase (FLuc) in complex with the infraluciferyl-adenylate analogue 5’-O-[(N-dehydroinfraluciferyl)-sulfamoyl] adenosine (iDLSA) resolved to a 3.2 Å resolution (PDB ID: 6HPS). (c) The structure of FLuc in complex with the iLH2 analogue iDLSA aligned to the reported structure of FLuc in complex with the LH2 analogue 5’-O-[(N-dehydroluciferyl)-sulfamoyl] adenosine (DLSA) (PDB ID: 4G36) (Sundlov et al., 2012) based on the FLuc N-terminal domain (residues 1–436). The structure of FLuc in complex with iLH2 is shown in red, and the structure of FLuc in complex with LH2 is shown in orange. (d) The structure of FLuc in complex with luciferin analogue DLSA (PDB ID: 4G36) (orange) and the infra-luciferin analogue iDLSA (red). Those residues within 4 Å of the substrate in each structure are highlighted in blue. (e) The table lists all residues within 4 Å of the both substrates, with those in orange or red only being found within 4 Å of DLSA and iDLSA respectively. (f) Highlights the same seven active site residues for both the aligned structures, with FLuc iLH2 residues in blue and FLuc LH2 residues in green. All analysis performed in PyMOL software (Schrodinger).