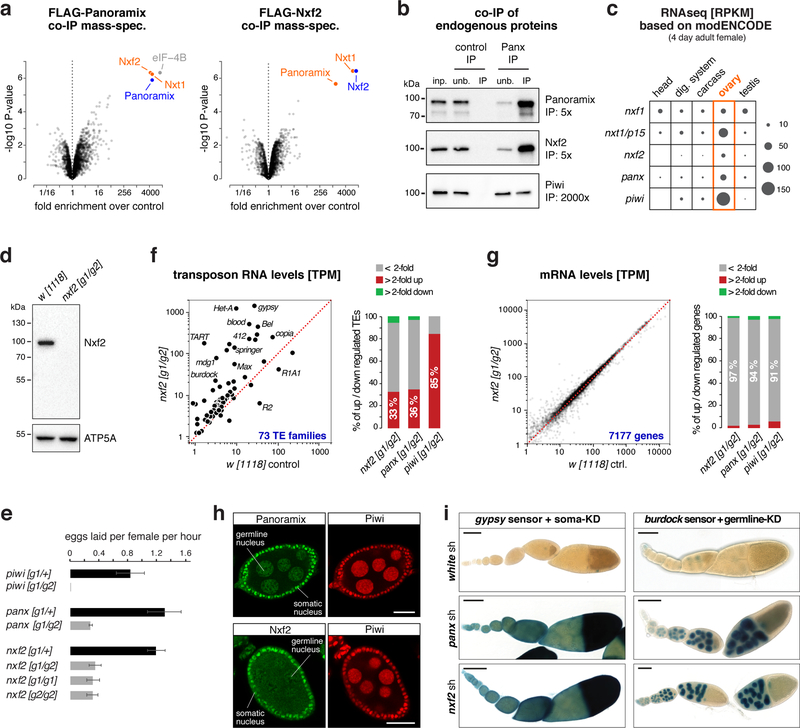
Figure 1. The Nxf2–Nxt1 heterodimer interacts with Panoramix.
a, Volcano plots showing fold enrichments of proteins determined by quantitative mass spectrometry in FLAG-Panoramix (left) or FLAG-Nxf2 (right) co-immunoprecipitates versus control (n=3 biological replicates). Experimental OSC lines stably expressed tagged Panoramix or Nxf2, wildtype OSCs served as control.
b, Western blot analysis of co-immunoprecipitation of endogenous Panoramix from nuclear OSC lysate. Relative amount loaded in immunoprecipitation (IP) compared to input (inp.) is indicated (unb. refers to unbound fraction; negative control: mouse IgGs).
c, Expression levels of indicated genes in indicated tissues based on modENCODE mRNA-seq data 31.
d, Western blot analysis of ovarian lysates with anti-Nxf2 antibody (ATP5A: loading control).
e, Average egg-laying rates of females with indicated genotype (n=3 biological replicates; error bars indicate s.d.). None of the eggs laid by panoramix or nxf2 mutant females developed into larvae.
f, g Left: Steady state ovarian RNA levels (transcripts per million) of transposons (f) and genes (g) in nxf2 mutants compared to control. Right: Number of differentially expressed genes or transposons in indicated genotypes.
h, Confocal images of wildtype egg chambers (scale bar: 20 μm) co-stained for Panoramix (top) or Nxf2 (bottom) with Piwi (for antibody specificity see Sup. Fig. 1f).
i, X-Gal stained ovarioles (scale bar: 0.1mm) from flies expressing gypsy-lacZ (soma sensor) or burdock-lacZ (germline sensor) plus indicated knockdown (KD) constructs.
Source data: panel a: Supplementary Table 1; panel f, g: Supplementary Table 2; uncropped blot images: Supplementary Data Set 1.