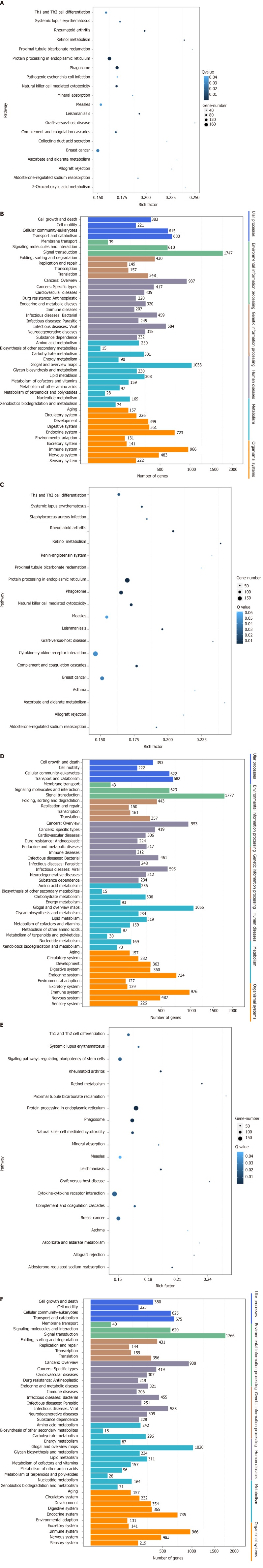
Figure 9.
Pathway enrichment analysis of gene targets of differentially expressed miRNAs. A: Comparison of the top 20 enriched pathway terms between vascular cell adhesion molecule-1 (VCAM-1) group and control group; B: Comparison of the Kyoto Encyclopedia of Genes and Genomes (KEGG) classification between VCAM-1 group and control group; C: Comparison of the top 20 enriched pathway terms between tumor necrosis factor (TNF) α group and control group; D: Comparison of the KEGG classification between TNFα group and control group; E: Comparison of the top 20 enriched pathway terms between interleukin 6 (IL6) group and control group; F: Comparison of the KEGG classification between IL6 group and control group. A, C, and E: The top 20 enriched pathway terms displayed as scatterplots. The rich factor is the ratio of target gene numbers annotated in this pathway term to all gene numbers annotated in this pathway term. The greater the rich factor, the greater the degree of enrichment. The Q-value is the corrected P-value and ranges from 0-1; the lower the Q-value, the greater the level of enrichment; B, D, and F: The X axis shows the number of target genes, and the Y axis shows the second KEGG pathway terms. The first pathway terms are indicated using different colors. The second pathway terms are subgroups of the first pathway terms and are grouped together on the X axis on the right side.